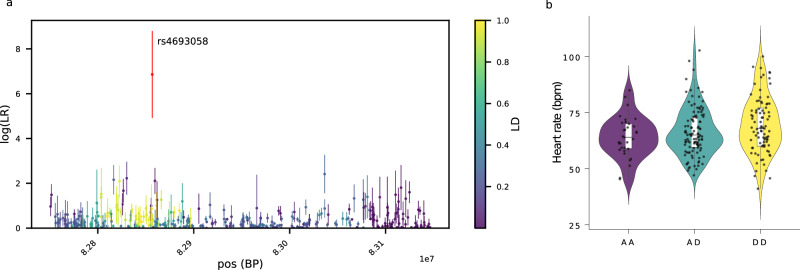
Fig. 4. Clues logLR and violin plots of the heart rate distribution depending on the genotype of the candidate SNP for the regions chr4:82750503-83146792 under selection in PNG lowlanders.
a logLR for SNPs in regions under selection after five runs of CLUES or 50 runs of CLUES for each of the five top SNPs in the candidate region. The candidate SNP rs4693058-C>T driving selection for the region is shown in red. The colour scale indicates linkage disequilibrium with the candidate SNP. CLUES logLR are presented as mean values +/− SD for n = 5 independent runs of CLUES (or n = 50 for the five top SNPs). b Violin plots of the heart rate distribution in PNG individuals (PNG highlanders, PNG lowlanders and PNG diversity set I, n = 232) depending on their genotype for the candidate SNP rs4693058-C>T (A = ancestral allele, D = derived allele under selection, AA = CC, AD = CT, DD = TT). The centre of the box-plot represents the median heart rate. The bounds of the box encompass the interquartile range (IQR = Q1–Q3), while the whiskers extend up to 1.5 × IQR units beyond the box boundaries. We used the ggplot2 package (v3.4.2) to generate the violin plots. Source data are provided as a Source Data file.