A phase II sequential chemoimmunotherapy trial in frontline stomach cancer demonstrates the importance of early tumor microenvironment remodeling to inform patients who benefit from adding the anti–PD-1 inhibitor pembrolizumab.
Abstract
Adding anti–programmed cell death protein 1 (anti–PD-1) to 5-fluorouracil (5-FU)/platinum improves survival in some advanced gastroesophageal adenocarcinomas (GEA). To understand the effects of chemotherapy and immunotherapy, we conducted a phase II first-line trial (n = 47) sequentially adding pembrolizumab to 5-FU/platinum in advanced GEA. Using serial biopsy of the primary tumor at baseline, after one cycle of 5-FU/platinum, and after the addition of pembrolizumab, we transcriptionally profiled 358,067 single cells to identify evolving multicellular tumor microenvironment (TME) networks. Chemotherapy induced early on-treatment multicellular hubs with tumor-reactive T-cell and M1-like macrophage interactions in slow progressors. Faster progression featured increased MUC5A and MSLN containing treatment resistance programs in tumor cells and M2-like macrophages with immunosuppressive stromal interactions. After pembrolizumab, we observed increased CD8 T-cell infiltration and development of an immunity hub involving tumor-reactive CXCL13 T-cell program and epithelial interferon-stimulated gene programs. Strategies to drive increases in antitumor immune hub formation could expand the portion of patients benefiting from anti–PD-1 approaches.
Significance:
The benefit of 5-FU/platinum with anti–PD-1 in first-line advanced gastric cancer is limited to patient subgroups. Using a trial with sequential anti–PD-1, we show coordinated induction of multicellular TME hubs informs the ability of anti–PD-1 to potentiate T cell–driven responses. Differential TME hub development highlights features that underlie clinical outcomes.
This article is featured in Selected Articles from This Issue, p. 695
INTRODUCTION
The first-line treatment of advanced gastroesophageal adenocarcinomas (GEA) has rapidly transitioned toward the broad adoption of concurrent chemoimmunotherapy. Unlike melanoma and non–small cell lung cancer (NSCLC) where a large portion of patients benefit from anti–PD-1 antibody monotherapy (aPD1), the activity of aPD1 in gastroesophageal adenocarcinomas is modest and heterogeneous (1, 2). Importantly, the optimal cooperativity between different treatment modalities (e.g., chemotherapy, immunotherapy, targeted agents) is understudied, and we are therefore limited in rationally designing novel combination treatments or sequencing strategies. This is in large part due to suboptimal preclinical model systems and clinical trial designs that use concurrent chemoimmunotherapy, thus making it difficult to dissect the relative contribution of individual modalities. To develop the next generation of gastroesophageal trial concepts, we need a deeper understanding of our current treatment paradigms.
To address this knowledge gap, we previously conducted a small pilot serial biopsy study in unselected metastatic gastric cancer patients treated with standard 5-FU/platinum (XELOX) chemotherapy and demonstrated early tumor microenvironment (TME) remodeling (3). Several features including on-treatment M1/M2-like macrophage repolarization and CD8+ T-cell influx were conserved among patients responding to standard 5-FU/oxaliplatin. Similar findings have been seen in some posttreatment surgical studies from nonmetastatic samples, highlighting a predictive and prognostic role for TME composition (4, 5). These findings support an ability of chemotherapy to generate a favorable environment in which to introduce immunomodulatory approaches, but were limited by lack of aPD1 incorporation, small sample size, and retrospective analyses.
The immune cells of the TME can assume a variety of phenotypes that can be defined by transcriptomic features and coexpression of multiple proteins. While modern spatial profiling technologies provide important insights into the spatial organization of the TME, substantial biology can be observed with TME reconstruction from bulk and single-cell RNA sequencing (scRNA-seq) data (6–8). Recently, covarying transcriptional programs in scRNA-seq data have been leveraged to predict multicellular interaction networks, or hubs, several of which have been spatially localized within tumors (9–13). While these studies have identified important insights into baseline tumor organization in microsatellite-high colorectal (MSI-H) patients, they have lacked on-treatment paired sampling to understand how these hubs may evolve under therapeutic pressures, and whether remodeling differs between patients who benefit and/or do not benefit from a given approach. We therefore designed a prospective single-arm sequential chemoimmunotherapy trial in patients with metastatic gastric cancer with serial sampling for deep correlative studies to build upon our prior observations. In the framework of this clinical trial, we sought to: (i) validate TME remodeling of 5-FU/platinum chemotherapy, (ii) understand the contribution of pembrolizumab on top of standard chemotherapy, and (iii) identify candidate pathways and cell types to nominate for further exploration in rational combination studies.
RESULTS
Clinical Activity and Molecular Characteristics
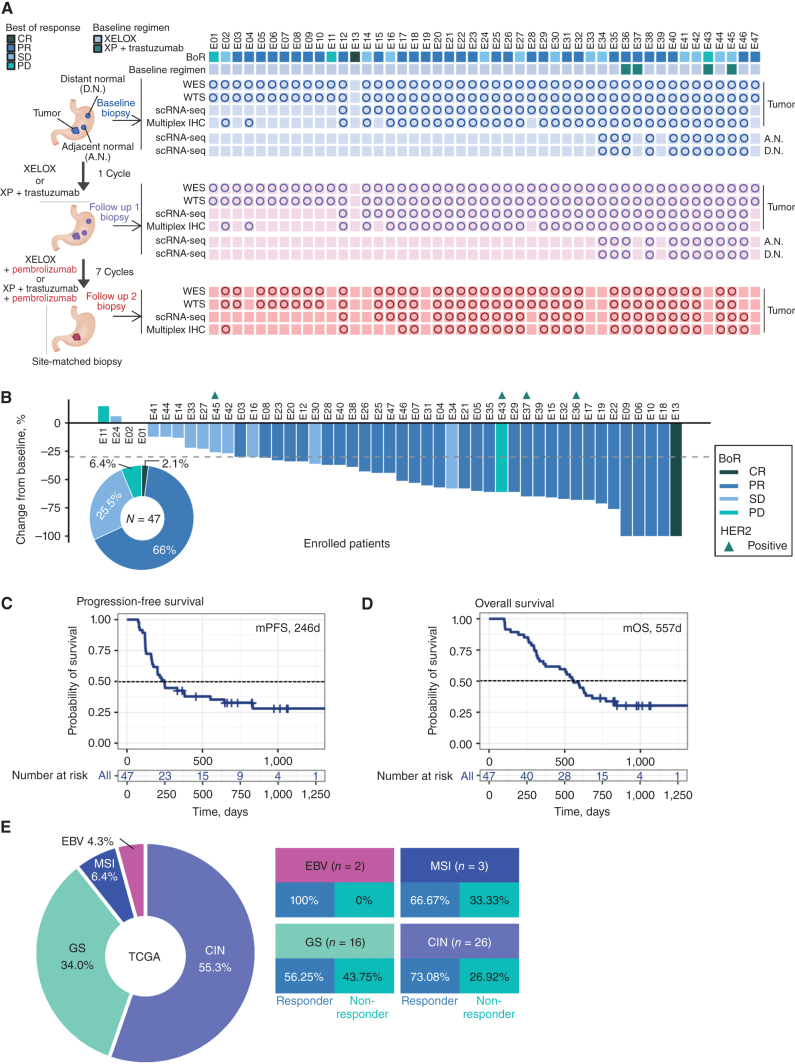
Trial accrual is completed, and 47 patients with treatment-naïve advanced gastric cancer (AGC) were enrolled between September 2020 and April 2022. The overall schema and sample collection strategy is outlined in Fig. 1A. Patients for whom no samples were available are not included in the correlative analyses. All tissue biopsies were taken from the primary tumor, and an attempt was done to serially biopsy the same region within the primary tumors using endoscopic mapping. The median age of the patients was 55 years (range 34–75 years), and most patients were men (80.9%). All patients were Korean. Forty-three patients (91.5%) received 5-FU/platinum chemotherapy and four (8.5%) received 5-FU/platinum chemotherapy with trastuzumab as first-line treatment per standard of care (Supplementary Table S1). Demographic features are shown in Supplementary Table S1. The primary objective was to characterize the TME changes, and the primary clinical endpoint was objective response rate by RECIST v1.1 (See Supplemental Trial Protocol). At the data cutoff on April 21, 2022, the overall response rate (ORR) was 68% (Fig. 1B; Supplementary Fig. S1A and S1B), and with a median follow up of 10.7 months the median progression-free survival (PFS) and overall survival (OS) was 8.2 months and 18.6 months, respectively (Fig. 1C and D; Supplementary Fig. S1C and S1D). The clinical activity is aligned with data from current phase III concurrent chemoimmunotherapy trials in Asian and Western patients (2, 14). As this trial contained only standard-of-care approaches the adverse events were in line with modern phase III GEA chemoimmunotherapy trials (Supplementary Table S2).
Figure 1.
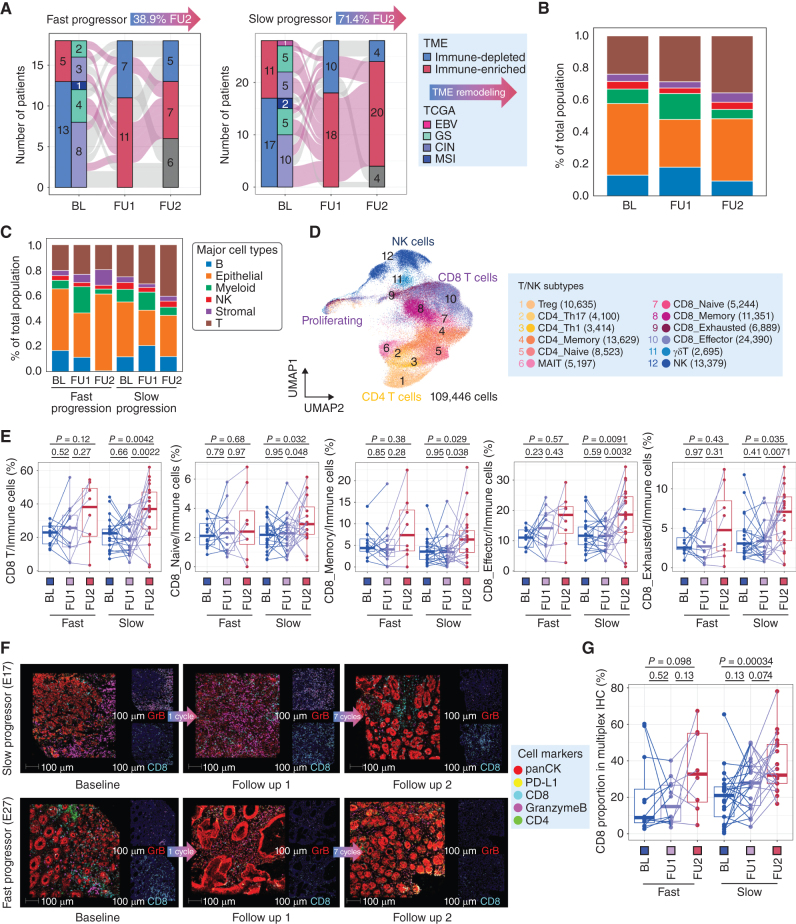
Phase II trial results and sample collection schema. A, Sample collection schedule and analysis platforms in a phase II sequential chemoimmunotherapy trial. Circles correspond to samples included in analyses. B, Waterfall plot demonstrating RECISTv1.1 response for patients in the trial. C, Clinical trial patient composition and response rates by TCGA molecular subgroup. D, Kaplan–Meier curve showing progression-free survival (PFS) by fast and slow progressor categorization. Statistical comparison performed using a log-rank test. E, Kaplan–Meier curve showing overall survival (OS) by fast and slow progressor categorization. Statistical comparison performed using a log-rank test.
The enrolled patient population is representative of the established genomic landscape described in the The Cancer Genome Atlas (TCGA) and Asian Cancer Research Group classifications (Fig. 1E; Supplementary Fig. S2; Supplementary Table S1; refs. 15, 16). Two patients had Epstein–Barr virus (EBV)-positive (EBV+) tumors (4.3%), three patients had tumors with microsatellite instability (MSI-high; 6.4%), 16 patients had genomically stable (GS) tumors (34%) and 26 patients had chromosomally instable (CIN) tumors (55%; Fig. 1E). Consistent with prior reports from our group, nearly all EBV+ or MSI-high patients achieved CR or PR (17, 18). Limited by low MSI-high and EBV+ patients, the median PFS by TCGA subgroup was 32.0 months for MSI-high, 20.4 months for EBV+, 7.6 months for CIN, and 9.4 months for GS tumors (Supplementary Table S1). Because RECIST stable disease (SD) can encompass heterogeneous clinical benefits (i.e., some SD patients achieve durable control), we conducted a post hoc analysis of patients by PFS, with fast progressors defined as PFS < 6 months (median PFS and OS was 121 and 271 days, respectively) and slow progressors as PFS > 6 months (median PFS and OS was 553 days and 842 days, respectively; ref. 19). The median PFS of 6 months was selected from modern phase III first-line trials and we observed a shorter overall survival in fast progressors (Supplementary Fig. S1C and S1D; refs. 14, 19–21). We, and others, have previously utilized this strategy to observe biology in single-arm trials involving immune agents where RECIST alone may underestimate patient benefit (22–24). There were no genomic features predictive of chemotherapy response, consistent with prior studies (3, 25). Tumor mutational burden was not significantly associated with clinical response in our cohort (Supplementary Fig. S2A and S2B). While this is the first trial to report outcomes by TCGA subtype the main intention was not to reexamine the clinical activity of standard approaches, but to generate a uniquely characterized cohort from which to better understand how our current therapies influence tumor, immune, and stromal features.
A Single Dose of 5-FU/Platinum Remodels Cell Type Composition in the TME
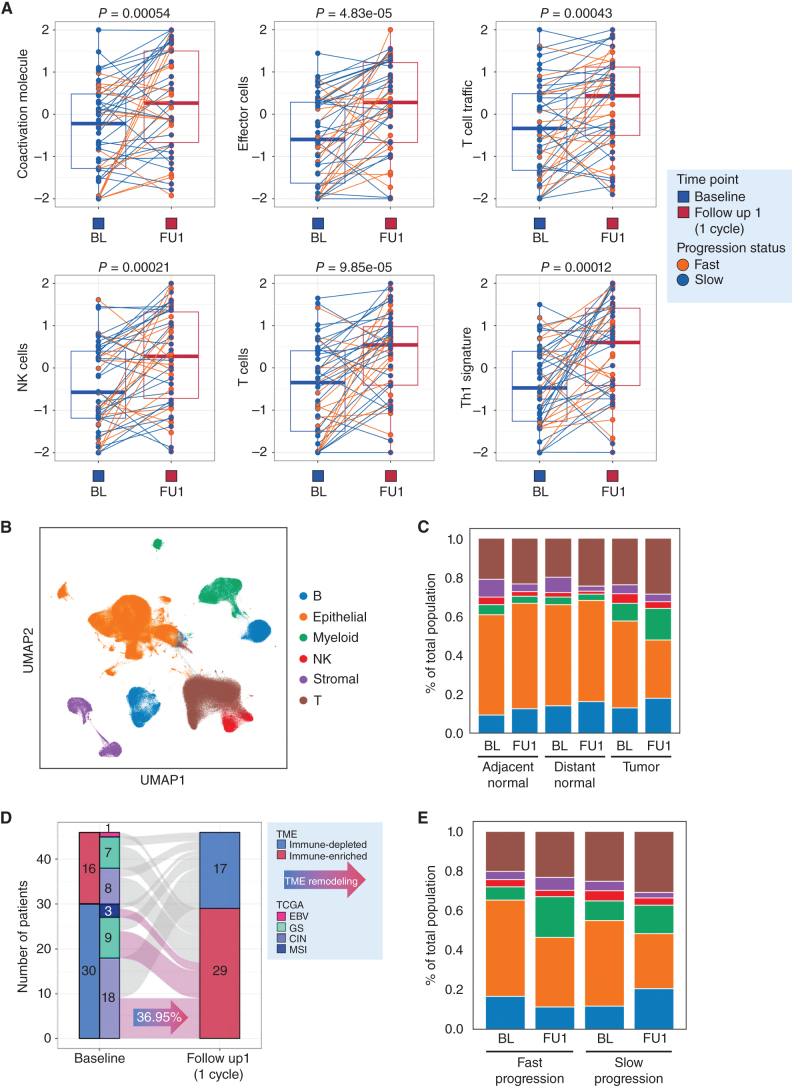
As we observed a lack of genomic chemotherapy response predictors, we sought to broadly understand the effects of standard platinum/5-FU on the tumor immune microenvironment. Our prior pilot work had suggested that 5-FU/platinum can lead to increased CD8+ T-cell infiltration and can reprogram macrophage populations (3). Comparing baseline (BL) and follow up 1 (after 1 cycle of chemotherapy; FU1), bulk transcriptomic analyses demonstrated broad upregulation of T-cell trafficking pathways, NK, and T-cell populations and Th1 signatures associated with IFNγ production (Fig. 2A; Supplementary Fig. S3A and S3B). Gene sets related to regulatory cells [including regulatory T cells (Treg), checkpoint inhibition and tumor-associated macrophages (TAM)] were also significantly higher at FU1 though the magnitude of change was less (Supplementary Fig. S3C). We additionally observed on-treatment enrichment of pathways involved in checkpoint inhibition and protumor cytokine signaling by gene-set variation analysis (GSVA; Supplementary Fig. S3B and S3C). To confirm tumor level compositional changes after chemo, we performed multiplex IHC (mIHC; Supplementary Figs. S3D, S4A, S4B, and S5). Consistent with bulk transcriptome data, we observed an increase in CD8+ lymphocyte infiltration during chemotherapy (Supplementary Figs. S3C, S4A, S4B, and S5). Without informing our analyses by clinical categories (responder vs. nonresponder or fast vs. slow progressor) our data confirms bulk immune cell compositional changes after a single cycle of standard 5-FU/oxaliplatin chemotherapy.
Figure 2.
A single cycle of 5-FU/platinum remodels the TME in advanced gastric cancer. A, Changes in enrichment in bulk RNA-seq data of immune-related pathways from baseline (Base) to FU1. Statistical comparison performed using a Wilcoxon signed-rank test. B, UMAP embedding of single-cell transcriptomes obtained from all samples in this trial. Labeled are canonical cell types. C, Cell-type proportions, obtained from scRNAseq data, in adjacent normal, distance normal and tumor tissue, at baseline (BL) and after 1 cycle of chemotherapy (FU1). D, Redistribution of TME subtypes following one cycle of 5FU/platinum chemotherapy. TME subtypes were obtained using a classification performed on bulk RNA-seq data. E, Cell type proportions, obtained from scRNAseq data, in tumor samples of fast and slow progressing patients at BL and FU1.
We then moved to identify granular cell types, substates and gene programs that underlie treatment response and resistance by leveraging our paired scRNA-seq data from BL and FU1. We examined single-cell gene expression profiles of 138 samples [BL tumor (n = 33), BL normal (adjacent normal, n = 11; distant normal, n = 11), FU1 tumor (n = 33), FU1 normal (adjacent normal, n = 11; distant normal, n = 11), and FU2 tumor (n = 28)]. After filtering low-quality cells, we collected a total of 358,067 cells. We performed unsupervised clustering and identified six major cell types using canonical marker genes, including epithelial cells, stromal cells, and immune cells (myeloid, T lymphoid, NK cells, B lymphoid; Fig. 2B; Supplementary Fig. S6A–S6F; refs. 3, 18, 26–28). Epithelial cells were categorized as tumor or normal cells using known marker genes of tumor cells and inferred copy-number variant (CNV) profiles with two methodologies, inferCNV (29) and Numbat (30; Supplementary Figs. S7A and S7B and S8). Supportive of the inferCNV classification we found in the 4 patients diagnosed with HER2+ disease by IHC, there was clear, yet heterogeneous amplification predicted at the HER2 locus on chromosome 17; interestingly, we found smaller subclonal populations with putative HER2 amplification in a subset of patients found to have HER2− disease (Supplementary Fig. S8).
Consistent with tumor cell death, we observed a contraction of the epithelial tumor component and expansion of myeloid and T-cell compartments after one cycle of chemotherapy without significant changes in adjacent or distant normal samples (Fig. 2C; Supplementary Figs. S6E and S6F and S9A; Supplementary Table S3). This trend was maintained when focusing on only GS and CIN patient samples as well (Supplementary Figs. S6F and S9B). TME archetypes are an important framework to understand shared immune cell features and have prognostic relevance (31). To understand archetype evolution, we classified all tumor samples into two distinct TME subtypes, immune-depleted or immune-enriched, using published TME subtypes associated with aPD1 outcomes (32). Seventeen patients (36.95%) converted from immune-depleted subtype to immune-enriched subtype after one cycle of chemotherapy treatment consistent with favorable TME remodeling (Fig. 2D). Although limited by sample size, we observed immune enriched and immune depleted subtypes across all TCGA subgroups, highlighting a complementary role for transcriptional classification (Fig. 2E; Supplementary Table S1). Interestingly, when stratified by fast versus slow progressors we noted a trend towards greater expansion of myeloid and stromal cells in fast progressors, and greater expansion of T and B cells in slow progressors suggesting even a single dose of chemotherapy may create a favorable TME trajectory in some patients, aligned with known predictors of IO benefit (Fig. 2E; ref. 33). As 4 patients received trastuzumab, we repeated the analysis after holding back the HER2+ patients and observed no significant differences (Supplementary Fig. S6E and S6F).
5-FU/Platinum Globally Alters Epithelial, Stromal, and Inflammatory Programs
To understand the gene expression programs induced within each cell type by a single dose of chemotherapy, we performed consensus nonnegative matrix factorization (cNMF) on each cell type at each timepoint individually, and identified between 9 and 48 programs for each cell type at each timepoint (Supplementary Figs. S10A–S10C, S11A–S11F, S12A–S12C, and Supplementary Table S4; refs. 13, 34, 35). To enhance the confidence in our cNMF observations, we performed 5-fold cross validation at each timepoint for each cell type and identified programs that were robust to missing cells across the dataset. To identify programs that were shared across timepoints for any given cell type, we performed a hypergeometric test of the top 100 weighted genes for all pairs of programs, yielding a subset of programs conserved across treatment conditions and another subset that was unique to each treatment condition (Supplementary Table S4). Among the shared programs across timepoints, we found several specific to particular immune or stromal cell subsets, such as a myofibroblast program (pS2_B, pS5_FU1), a Treg program (pT12_B, pT3_FU1, pT6_FU2), a myeloid regulatory dendritic cell program (mregDC; pM8_B, pM17_FU1, pM20_FU2), and a plasmacytoid dendritic cell program (pDC; pM6_B, pM4_FU1, pM8_FU2), among others (Supplementary Figs. S11A–S11F and S12A–S12C). Several shared programs were expressed across cell subtypes, such as stromal MHCII (pS1_B, pS11_FU1, pS17_FU2) and proliferation programs (pS3_B, pS9_FU1, pS9_FU2), myeloid inflammatory, IFN-stimulated gene (ISG), and IL1 programs (pM2_B, pM10_FU1, pM13_FU1, pM13_FU2, pM14_FU2), and T-cell translation (pT1_B, pT10_FU1), HSP (pT4_B, pT12_FU1, pT8_FU2), NFkB/JUN/FOS (pT11_B, pT15_FU1, pT3_FU2), and MHCII, IFNγ/ISG and activation programs (pT14_B, pT9_FU1, pT13_FU1, pT14_FU1, pT11_FU2; Supplementary Figs. S11A–S11F and S12A–S12C).
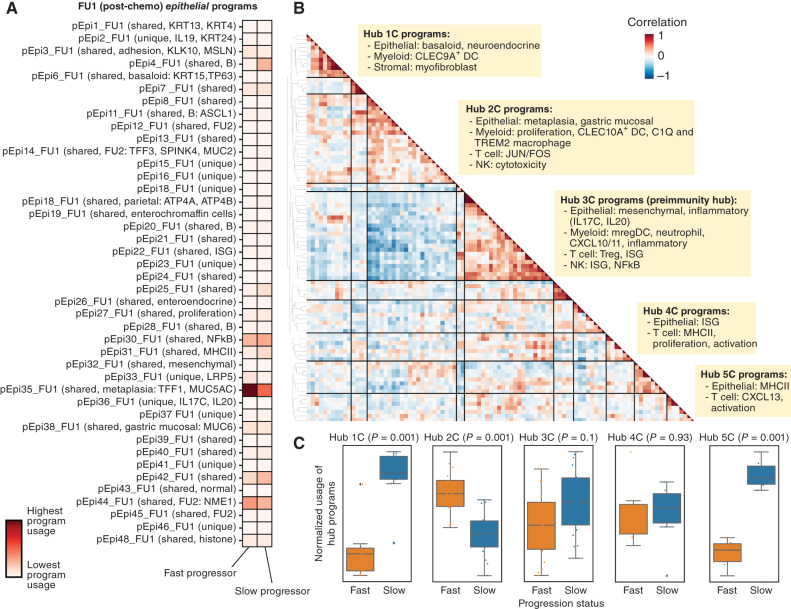
Importantly, we found a subset of shared epithelial programs across many patients at each timepoint (e.g., pEpi3_B, among others), which included basaloid (pEpi7_B, pEpi6_FU1), parietal (pEpi9_B), proliferation (pEpi10_B), KRT20+ (pEpi16_B), gastric mucosal/MHCII (pEpi17_B), and adhesion programs (pEpi19_B; Supplementary Fig. S10A–S10C). We found epithelial mesothelin (pEpi3_FU1), ISG (pEpi22_FU1), and metaplasia (pEpi35_FU1) programs that were present only after chemotherapy treatment, suggesting these may reflect adaptive epithelial responses to treatment. Other programs, such as a neuronal-like (pEpi13_B, pEpi11_FU1) program marked by ASCL1 expression, were present across timepoints but were largely sample-specific. We next sought to identify programs that were differentially induced in epithelial cells when comparing fast and slow progressing patients (Fig. 3A; Supplementary Figs. S13A and S13B and S14). We found that the metaplasia program, marked by TFF1 (36) and MUC5AC (37) as top weighted genes, and the adhesion program, marked by MSLN as a top weighted gene, had significantly higher usages in epithelial cells postchemotherapy (FU1) in fast progressors compared with slow progressors (Fig. 3A; Supplementary Fig. S13A). This potentially implicates chemotherapy-induced metaplastic programs as a poor prognostic marker and this difference in metaplasia program usage between fast and slow progressing patients was maintained after immunotherapy (Supplementary Fig. S13B). Recently, MUC5AC was observed to be enriched among colorectal cancer patients with peritoneal disease, a particularly poor prognosis and fast progressing clinical subgroup (38). Similarly, MSLN expression is a poor prognostic feature and an active area of investigation for cellular therapies (39, 40).
Figure 3.
Identification of covarying gene programs that underlie chemotherapy resistance and response. A, cNMF was performed on epithelial cells at FU1. Shown is the mean usage of each cNMF gene program in the epithelial cells of fast and slow progressing patients. B, Heat map showing pairwise correlation of gene program activities across all patient samples at FU1 using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK, and stromal cells. Hierarchical clustering was performed to identify clusters of covarying proteins, which have been labeled as Hub1C to 5C. C, Average z-scored usage of all gene programs in each hub split by fast and slow progressing patients. Statistical comparison performed using a two-sample t test with Bonferroni correction.
5-FU/Platinum Remodels Cellular Interaction Networks and Drives Multicellular Hubs in the TME
We next sought to identify cellular programs across all cell types that may influence each other after a single dose of chemotherapy. Toward this end, we used a methodology to identify groups of cell type–specific programs that covaried across all samples at a given time point. For each sample, we quantified the program usage of each program within the cell type it was identified, focusing on epithelial, stromal, myeloid, T, and NK cells. We subsequently looked at the correlation of these program usages over all samples and identified groups of covarying programs (termed “multicellular hubs”; Supplementary Figs. S15–S17; Supplementary Table S5). We found several multicellular hubs at baseline (Supplementary Fig. S15) and after chemotherapy (Fig. 3B; Supplementary Fig. S16). Most notably, we found five major hubs after chemotherapy each with distinct properties (Fig. 3B). This included: (i) covarying epithelial basaloid and myofibroblast programs (Hub 1C); (ii) epithelial metaplasia, myeloid proliferation and C1Q and TREM2 macrophage programs (Hub 2C); (3) epithelial mesenchymal and inflammatory, mregDC, myeloid CXCL10/11, Treg and T-cell ISG programs (Hub 3C); (iv) epithelial ISG and T-cell activation programs (Hub 4C); and (v) epithelial MHCII and T-cell CXCL13 and activation programs (Hub 5C).
Hubs 3C and 5C, while distinct at this time point, appeared to collectively include the components of a recently described multicellular immunity hub (10) that includes a positive feedback loop between CXCL10/11 expressing myeloid and tumor cells, and ISG expressing and tumor-reactive CXCL13+ T-cell subsets (10, 41, 42). These results suggest that a single cycle of standard 5-FU/platinum may steer the formation of key immunity hubs in GEA. We therefore hypothesized that the composition and degree to which antitumor immunity hubs are induced by chemotherapy may inform the ability of additional ICB to induce a durable antitumor response and durable clinical benefit. We next assessed the relative representation of each hub in slow versus fast progressors (note: hubs were discovered independent of response status and only by looking at covariation across all samples); Hubs 1C and 5C programs had significantly higher usage in slow progressors compared with fast progressors, and hub 3C also trended in that direction, (Fig. 3C), consistent with proposed role of Hubs 3C and 5C in portending favorable immune response. Importantly, Hub 2C programs had higher usage in fast progressors compared with slow progressors (Fig. 3C). As Hub 2C included covariation of the poorly prognostic metaplasia program with several macrophage programs, we opted to further probe the role of chemotherapy treatment in altering macrophage subsets (Fig. 3B; Supplementary Fig. S11A–S11C).
5-FU/Platinum Reprograms Macrophages Subsets
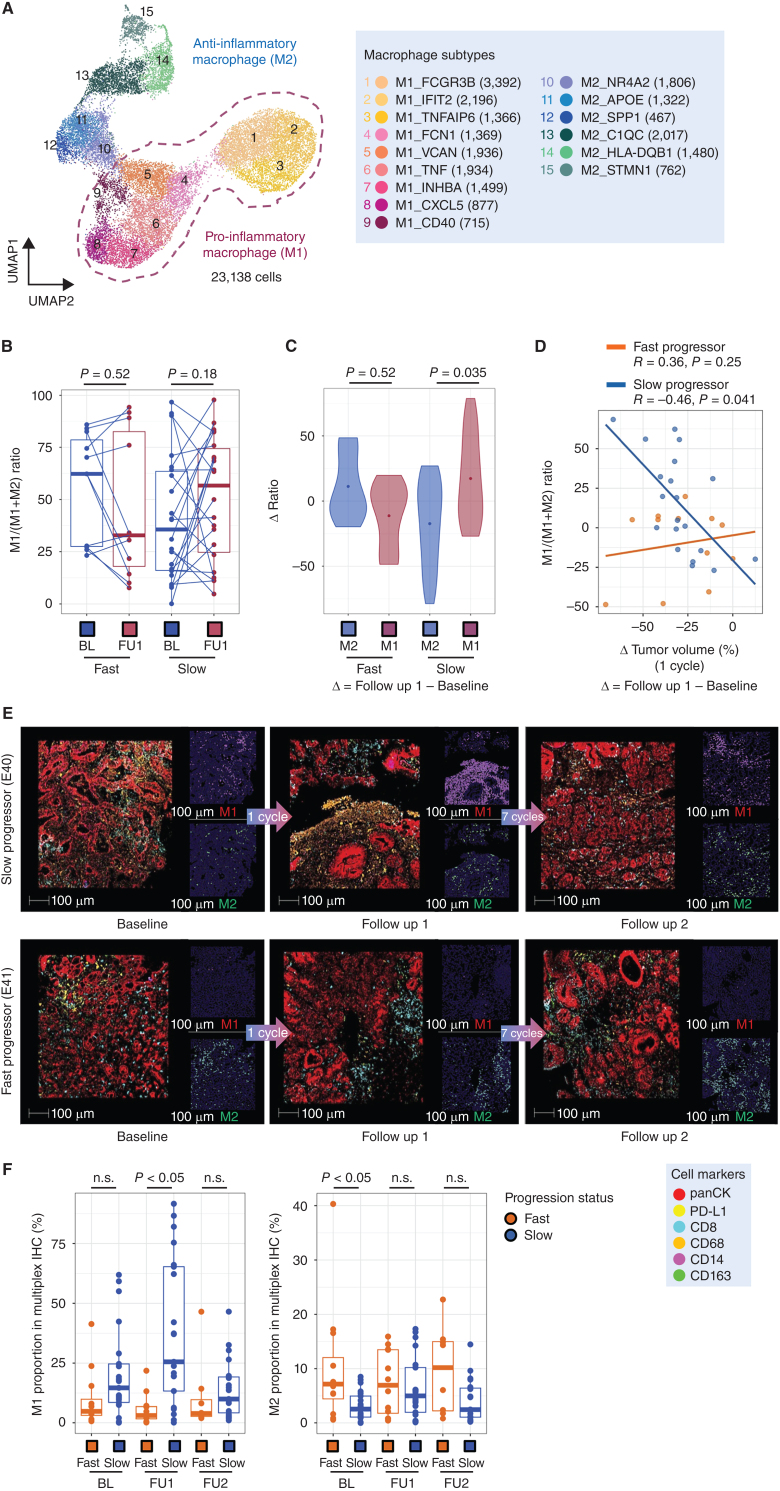
Previously, we had observed a role for early on-treatment TAM orientation in identifying clinical responders in GEA, and a relationship between TAMs and T-cell exhaustion programs in cancer is well described, particularly in the inner regions of tumors (43, 44). Although overly simplistic, the M1 versus M2 framework is useful for conceptualizing TAM populations, and M2-like TAMs can impair antigen presentation and limit T-cell activation (43, 45, 46). In this context, we explored TAM associations with response. We first performed a high-level clustering of all myeloid cells, identifying dendritic cell, monocyte/macrophage, and mast cell populations (Supplementary Figs. S18A and S18B). To further refine TAM subsets, we performed subclustering of the TAM population and identified cells divided among 15 subtype clusters (Fig. 4A; Supplementary Fig. S19A and S19B). To better dichotomize the TAMs, we grouped them into proinflammatory macrophage subsets (M1: M1_FCGR3B, M1_IFIT2, M1_TNFAIP6, M1_FCN1, M1_VCAN, M1_TNF, M1_INHBA, M1_CXCL5, M1_CD40) and anti-inflammatory subsets (M2: M2_NR4A2, M2_APOE, M2_SPP1, M2_C1QC, M2_TMEM176B, M2_HLA-DQB1, M2_STMN1). Interestingly, we found that the C1Q and TREM2 macrophage programs that covaried with the poorly prognostic epithelial metaplasia program were most highly expressed in M2 macrophage subsets (Supplementary Fig. S11B), consistent the proposed negative prognostic role of M2-like macrophages. As has been seen in prior work, we found proinflammatory genes, such as IL1B and S100A8 were significantly upregulated in M1 clusters (Supplementary Fig. S19B).
Figure 4.
Chemotherapy leads to macrophage repolarization in patients with favorable responses. A, UMAP embedding of single-cell transcriptomes of all macrophages from all samples in this trial. Labeled are granular macrophage subtypes, including designation of M1 and M2 subtypes. B, Relative proportion of M1 macrophages of all macrophages, obtained from scRNAseq data, at BL and FU1 in fast and slow progressing patients. Statistical comparison performed using a Wilcoxon signed-rank test. C, Change in M1 and M2 macrophage proportions from BL to FU1, obtained from scRNAseq data, in fast and slow progressing patients. Statistical comparison performed using a Wilcoxon signed-rank test. D, Change in relative M1 proportion from BL to FU1 plotted against change in tumor volume after 1 cycle of chemotherapy, segregated by fast and slow progressing patients. E, Multiplexed immunofluorescence (mIF) images of BL, FU1 and FU2 samples from two patients, E40 (slow progressor) and E41 (fast progressor), staining for panCK, PD-L1, CD163, CD68, CD14 and CD8. F, Proportion of M1 macrophages, obtained from mIF images, at BL and FU1 in fast versus slow progressing patients. Statistical comparison performed using a Wilcoxon signed-rank test.
Using the M1/M2 conceptual structure we compared TAMs from tumor tissues at baseline and FU1. We observed divergent patterns of M1-like proportions between fast and slow progressor groups (Supplementary Fig. S19C and S19D). In a prior pilot study, we noted the relevance of the M1/M1+M2 ratio in segregating responders (3, 18). In this prospective trial we confirmed changes in the M1/(M1+M2) ratio with a trend for increased M1/(M1+M2) ratio and significantly greater magnitude of change in the slow progressor group, and this was consistent when doing the comparison between clinically defined RECIST responders and nonresponders (Fig. 4B and C; Supplementary Fig. S19C–S19H). Within the M2-like subset SPP1 expression is associated with proangiogenic signaling and worse outcomes with immunotherapy (47, 48). We examined M2-SPP1 expression and demonstrated a decrease in M2-SPP1 proportions only in responders (Supplementary Fig. S19F). To highlight the clinical relevance of early on-treatment M1/(M1+M2) ratio changes, we examined the relationship to CT and endoscopic images. Patients with increased M1/(M1+M2) ratio showed decreased tumor volume by CT scan images, PET scan image and inflammation in the stomach by endoscopic images (Fig. 4D; Supplementary Figs. S19G and S20A). However, patients with low on-treatment M1/(M1+M2) ratio after one cycle of chemotherapy showed a limited response on CT scan, PET scan, and endoscopy (Supplementary Fig. S20B). Given that the expansion of M1-like TAMs in tissues was associated with a clinical response, we sought to confirm our scRNA-seq findings with mIHC. Using established macrophage lineage markers (CD14, CD68, CD163), we first confirmed early M1 expansion on treatment with mIHC (Supplementary Fig. S5). Using the mIHC-determined M1 proportion from 33 patients there was significant expansion during chemotherapy in the slow progressor group aligning with our scRNAseq data (Fig. 4E and F; Supplementary Fig. S20C). Overall, early on-treatment changes in macrophage subsets predicted subsequent clinical outcomes.
Stromal Cell and Macrophage Interactions Confer an Unfavorable TME
Having confirmed the clinical relevance of macrophage subset composition, we wanted to more deeply understand M2 interactions with other TME components. Interestingly, we found that the four patients with increased stromal cell proportion (E24, E27, E30, and E33) were all fast progressors and had a larger change in proportion of M2-like macrophages after one cycle of chemotherapy (Supplementary Fig. S21A and S21B). We next performed a subclustering of stromal subsets and identified five distinct clusters of cells: fibroblasts, myofibroblasts, endothelial cells, pericytes, and glial cells (Supplementary Fig. S21C and S21D). We subsequently noted that myofibroblasts are decreased in number in slow progressors at FU1, but not in fast progressors (Supplementary Figs. S21E). To further investigate how M2-like macrophages might influence stromal components, and vice versa, we scored M2 macrophage subsets on signatures for phagocytosis, angiogenesis, and the PDGF pathway, which is known to influence fibroblast behavior. We found APOE and C1QC macrophages to have the highest phagocytosis scores, SPP1 macrophages to have the highest angiogenesis scores, and finally for NR4A2, APOE and SPP1 macrophages to have the highest PDGF pathway scores (Supplementary Figs. S22).
To better delineate the specific cellular interactions that may underlie cell–cell interactions after chemotherapy, we applied a methodology to infer ligand–receptor pairs (CellphoneDB; ref. 49) between cell types using scRNAseq data and applied it to each treatment time point and to normal tissues separately (Supplementary Table S6). We found that the number of predicted interactions were highest for stromal cells with other stromal cells or with myeloid cells (Supplementary Fig. S23A–S23D). We homed in on predicted L–R interactions within tumor tissues that were distinct from normal tissue (Supplementary Table S7), and additionally were unique to FU1 samples compared with baseline samples (Supplementary Table S7). We found that after chemotherapy, LGALS9 and SIRPa on myeloid cells served as ligands to CD47 on stromal cells, thus inhibiting phagocytosis by myeloid cells (50). In addition, we found several other myeloid–stromal ligand–receptor interactions that may influence macrophage polarization, including stromal cell produced E-selectin and macrophage inhibitory factor (MIF). Importantly, we found several ligands in myeloid cells shared at the baseline timepoint that may influence stromal behavior, including CD55, IL8, TNF, TGFB1, and SPP1.
The Addition of Pembrolizumab Enhances T-cell Antitumor Immune Remodeling
A main goal of the trial design was to understand early chemotherapy-driven TME features and their impact on aPD1 benefit. We first applied a validated gene expression signature predictive of pembrolizumab benefit and demonstrated an increase with chemotherapy alone that was furthered by the addition of pembrolizumab across all patients (Supplementary Fig. S24A; refs. 51–53). After a single cycle of 5-FU/oxaliplatin, we observed increased expression of representative genes of immunogenic cell death (ICD) in the slow progressor group, but not the fast progressor group (Supplementary Fig. S24B; ref. 54). Consistent with these findings, we found a larger proportion of slow progressor patients had remodeled TMEs toward immune enriched environments compared with fast progressors (Fig. 5A). In our serial scRNAseq data, the composition of the TME was altered after aPD1 with an increased proportion of T cells after chemotherapy treatment (Fig. 5B). When comparing fast and slow progressors, we found that the expansion of T cells after aPD1 was predominantly in slow progressors and was not observed in fast progressors (Fig. 5C; Supplementary Fig. S9B). This suggests a working model whereby the early chemotherapy-driven TME remodeling leading to the formation of an immunity hub containing CXCL13+ T cells is a central determinant of whether or not the addition of aPD1 can potentiate an antitumor T-cell response. We note, however, given the sequential nature of treatments given in the trial, we cannot exclude the possibility that the changes observed over time in slow progressors may be independent of immunotherapy treatment. That is, early TME modeling may serve as a prognostic marker as opposed to having predictive value for aPD1 benefit.
Figure 5.
The addition of immunotherapy to 5-FU/platinum chemotherapy redistributes T-cell phenotypes. A, Remodeling of TME from immune depleted to immune-enriched environments derived from bulk RNA-seq profiles in fast and slow progressing patients, shown across timepoints. B, Cell type proportions, obtained from scRNAseq data, of all tumor samples at BL, FU1 and after immunotherapy treatment (FU2). C, Cell type proportions, obtained from scRNAseq data, in tumor samples of fast and slow progressing patients at BL, FU1, and FU2. D, UMAP embedding of single-cell transcriptomes of all T and NK cells from all samples in this trial. Labeled are granular T- and NK-cell subtypes. E, Cell type proportions as a proportion of all immune cells, obtained from scRNAseq data, of total, naïve, memory, effector, and exhausted CD8 T cells. Statistical comparisons performed using a Wilcoxon signed-rank test. F, Multiplexed immunofluorescence (mIF) images of BL, FU1 and FU2 samples from two patients, E17 (slow progressor) and E27 (fast progressor), staining for panCK, PD-L1, CD4, CD8, and Granzyme B. G, Proportion of CD8 T cells macrophages, obtained from mIF images, at BL, FU1 and FU2 in fast versus slow progressing patients. Statistical comparison performed using a Wilcoxon signed-rank test.
To understand which T-cell subsets were most strongly affected by aPD1 treatment, we next subclustered the T and NK cells in our scRNAseq dataset to identify more granular subtypes (Fig. 5D; Supplementary Fig. S25A and S25B). We found significant CD8 T-cell expansion across timepoints (B, FU1, FU2) in slow progressor patients but not in fast progressor patients (Fig. 5E), and this was consistent when comparing responders to nonresponders (Supplementary Fig. S26A). We validated that there was increased CD8 T-cell expansion in slow progressors (and treatment responders) using mIF for CD4, CD8, GzmB, and PD-L1 (Fig. 5F and G; Supplementary Figs. S5 and S26B and S26C). Among CD8 T-cell subsets, we found that CD8 naïve, memory and exhausted subsets were significantly expanded after aPD1 in slow progressors, but not in fast progressors (Fig. 5E; Supplementary Fig. S26D). CD8 effector subsets were expanded in both slow and fast progressors after aPD1 (Fig. 5E); however, when performing the analysis comparing responder and nonresponder patients, this only held true in the responder subgroup (Supplementary Fig. S26D). Of note, within the CD4 T-cell compartment, we found that Tregs and Th1 cells were most altered in frequency across treatment timepoints (Supplementary Fig. S26E).
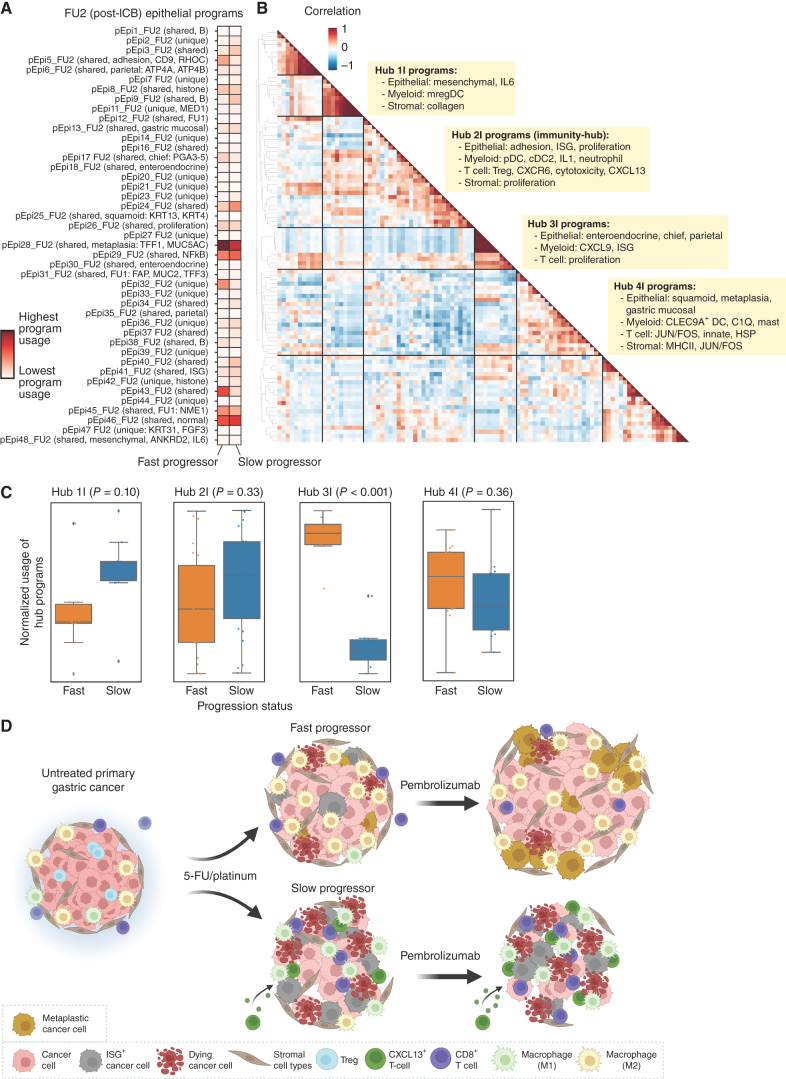
In our analysis of gene expression programs, we had identified a tumor-reactive T-cell program, which included CXCL13 as a top weighted gene, that was only identified postchemotherapy (pT18_FU1) and ICB (pT12_FU2; Fig. 3A; Supplementary Fig. S11F). We found that the usage of these programs was higher in T cells of slow progressors compared with fast progressors (Fig. 3A; Supplementary Figs. S26F and S27A and S27B), consistent with literature reports that have identified CXCL13+ CD8 T cells as a tumor-reactive T-cell subset (41, 42, 48). We further validated that CXCL13+ was increased in single CD8 T cells across timepoints, in addition to other tumor-reactive (CD39, CD103) and costimulatory markers (4–1BB, GITR; Supplementary Fig. S27C). To further investigate the evolution of covarying hubs identified after chemotherapy treatment (Fig. 3B), we next looked for covarying gene programs across samples after immunotherapy treatment (Fig. 6A and B). We identified 4 major hubs postimmunotherapy, and notably found that the CXCL13 T-cell program covaried with epithelial adhesion, ISG and proliferation programs, myeloid dendritic cell and IL1 programs, Treg and T-cell cytotoxicity programs, and stromal proliferation programs (Hub 2I; Fig. 6B; Supplementary Fig. S17). We termed this the “immunity hub” and note that we find that compared to chemotherapy treatment alone, the addition of immunotherapy led to the covariation of the tumor-reactive CXCL13 program with other programs that have several similar features to those seen in other multicellular hubs identified in colorectal cancer (10). This includes the presence of epithelial ISG and Treg programs, but also with distinct properties from the hubs identified in colorectal cancer that include the presence of myeloid IL1 and dendritic cell programs.
Figure 6.
Multicellular hubs underlie chemoimmunotherapy resistance and response. A, cNMF was performed on epithelial cells at FU2. Shown is the mean usage of each cNMF gene program in the epithelial cells of fast and slow progressing patients. B, Heat map showing pairwise correlation of gene program activities across all patient samples at FU2 using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK, and stromal cells. Hierarchical clustering was performed to identify clusters of covarying proteins, which have been labeled as Hub1C to 5C. C, Average z-scored usage of all gene programs in each hub split by fast and slow progressing patients. Statistical comparison performed using a two-sample t test with Bonferroni correction. D, Summary schematic of proposed changes to the TME after 1 cycle of chemotherapy and chemoimmunotherapy in fast versus slow progressing patients; in particular, fast progressing patients have induction of metaplasia programs and increased abundance of suppressive M2 macrophages. Slow progressing patients have increased infiltration of CXCL13+ CD8 T cells after chemotherapy, and increased tumor-intrinsic ISG induction and inflammatory M1 macrophage subsets.
We identified three other notable multicellular hubs in the aPD1 treated samples that include: (i) epithelial mesenchymal and IL6, mregDC, and stromal collagen programs (Hub 1I); (ii) epithelial neuroendocrine, chief and parietal, and myeloid CXCL9 and ISG programs (Hub 3I); and (iii) epithelial squamous and metaplasia, myeloid C1Q macrophage and CLEC9+ DC, T-cell JUN/FOS, innate and HSP, and stromal MHCII and JUN/FOS programs (Hub 4I). We next analyzed hub participation after aPD1 between slow and fast progressors, and we found that compared to fast progressors, slow progressors trended toward higher usage of programs in Hub 1I and Hub 2I, the immunity hub, and had significantly lower usage of 3I, both of which may therefore be features of aPD1 resistance (Fig. 6C).
Despite being more highly represented in slow progressors, Hub 1I centered around several immune features known to directly antagonize immunotherapy response most notably mregDCs (55) and IL6 (56). Notably, circulating plasma IL6 was higher among fast progressors in our trial (Supplementary Fig. S22). We suspect this paradox may likely be due to the source of IL6 in circulation and as reported in previous studies (bioRxiv 2022.02.02.478819). Whereas in Hub 1I, we find epithelial cells as a major source of IL6, several studies have reported that IL6 produced by other cell types, particularly myeloid cells, may be poorly prognostic (57). Hub 4I contained immunosuppressive C1Q+ TAM (58) programs covarying with markers known to be involved in T-cell exhaustion and impaired antitumor immunity (JUN/FOS; ref. 59). The epithelial programs covarying with the immunosuppressive immune cell programs may hint at epithelial cell plasticity and are enriched for squamoid and metaplasia (Hub 4I) programs, which are implicated in therapeutic resistance (60, 61).
DISCUSSION
Efforts to identify determinants of response to cytotoxic chemotherapy and immune checkpoint blockade, and dissecting the evolution of immune cell populations and interactions under therapeutic pressures has been largely limited to comparisons of pretreatment and temporally distant posttreatment and/or progression samples across solid tumors (11, 62, 63). While useful, this framework limits the ability to develop early, true on-treatment biologic insights and biomarkers to inform biologically adapted clinical trial concepts and nominate therapeutic targets. Here we present a novel and large sequential sampling cohort in the framework of a prospective phase II chemoimmunotherapy trial in AGC. To our knowledge, this is the first and largest trial of its kind to couple serial true on-treatment biopsies with multiparametric molecular characterization in advanced GEA.
Taken together our data point toward an early initial complex coordinated response in primary tumor lesions to cytotoxic chemotherapy that differs between patients who go on to develop response/benefit and those that do not (Fig. 6D). Notably, the baseline TME composition appears to have less impact than the early adaptive changes induced by chemotherapy. Using an initial window after 1 cycle of 5-FU/platinum (∼2–3 weeks between baseline; BL and follow up 1; FU1 samples) we uncover early on treatment patterns that converge on central components of the TME, specifically the myeloid and T-cell interactions with both the epithelial and stromal elements. This is largely consistent with the observations that presence, and in our case induction, of immune archetypes are important modulators of the ability to mount a T cell–driven immune response with aPD1 approaches. To gain a more granular insight into drivers of TME modulation, we leveraged covarying gene programs to infer multicellular interaction networks across our timepoints. This strategy has identified key conserved programs and “immune hubs” in colorectal cancer but was limited to untreated earlier stage patient samples (10, 34). How these networks evolve during therapeutic pressure has been a scientific blind spot. The described hubs with colocalization of IFNG+ and CXCL13+ T cell programs is something we observed, but only after treatment with chemotherapy (FU1). Interestingly, Pelka and colleagues did not observe this preexisting hub when restricting analysis to microsatellite stable (MSS) colorectal cancers (10, 64). Although limited by sample size, particularly for MSI-high and EBV+ subtypes, were observed TME evolution independent of genomic TCGA classification, highlighting the heterogeneity within the general TCGA subgroups.
Conceptually, a single dose of chemotherapy looks to incite a turf war between opposing communities (hubs), the prevailing composition and usage of which will influence the ability of aPD1 addition to further the antitumor immune response. This idea is reflected in the relative hub usage in fast and slow progressors with fast progressors demonstrating increase in epithelial metaplasia, myeloid proliferation, and C1Q and TREM2 macrophage programs (Hub 2C). Predicted cellular interactions in this group are associated with known adaptive resistance mechanisms including tumoral plasticity and IL6 (65–67). Conversely, we observe a portion of patients with suggestions of proimmunity hubs evolving after chemo and furthered by the addition of aPD1 and it is tempting to speculate that these patients are indeed some of the patients who derive very durable benefit, often described in the “tail of the curve” in aPD1 clinical trials. We in fact see greater representations of the aforementioned hubs after chemotherapy and after aPD1 in patients that are slow to progress (have more durable responses) compared with those that progress faster. From a clinical perspective, it remains unclear whether the optimal strategy is to attempt to restrain the initial formation of antiimmunity hubs with agents that deprive the TME of immunosuppressive signals as has been attempted with neutralizing antibodies to IL6 (56) and DKK1 (68), or to try to directly stimulate the induction of proimmunity hubs with agonist approaches like STING (69, 70) or TLR7/8 (71, 72). Toward the later approach, we find that programs for plasmacytoid and conventional dendritic cell subsets were present in hubs that favored slow progression (e.g., Hub 2I). More nonspecific strategies to remodel the TME such as combinations with aPD1, chemotherapy and VEGF small-molecule inhibitors are also under clinical investigation (73).
Although our dataset lacks high resolution spatial characterization, some of the broad features raised in our scRNAseq analyses are recapitulated in our multiplex IHC/IF analysis. To date, some of the correlations between inferred network structure from scRNAseq and ground truth analysis such as IHC and IF and emerging spatial transcriptomics appear to largely hold, which is reassuring for our data (10, 11, 43, 63, 64, 74, 75). As we needed to prioritize patient safety, our serial samples are limited to endoscopic biopsies of the primary tumor, and we recognize the potential value for complementary spatial transcriptomics approaches. For instance, we nominate an increase in cDC1 programs that parallels CD8 effector increase and M2 decrease only in responders. Receptor–ligand predictions point to a direct communication among these cell types, but we cannot confirm the spatial orientation. In a recent analysis from a PD-1 containing CRC trial (Keynote-177) the spatial proximity between CD74+ macrophages and PD1+ CD8 T cells was a robust response predictor (76).
TAM, particularly the alternatively activated (M2-like) subset are known to facilitate tumor-promoting processes including angiogenesis, impede antigen presentation, and suppress the tumoricidal functions of CD8 T cells and NK cells (77–80). Recent data suggest SPP1-high TAMs are linked to EMT, higher angiogenesis scores, and liver metastasis (47, 81, 82). We observed a decrease in SPP1 expression only among responding patients, and a suggestion of increased macrophage-derived MIF inhibiting T-cell proliferation when examining M2-like macrophage interactions with T cells. MIF is known to directly antagonize immune responses in melanoma (83). Despite preclinical rationale for repolarizing M2-like TAMs toward the more antitumor M1-like state, the clinical development has fallen short in several chemotherapy-free approaches, including in GEA (reviewed in ref. 44). Additional support for our broad observations is seen in WTS analyses of samples from the phase III frontline CheckMate-649 trial and adjuvant CheckMate-577 trials where increased M2 signature and high stromal signatures were associated with lesser benefit from PD-1.
The inciting signals, or mix thereof, that drives the initial formation of pro- and anti-immunity hubs is an area of important ongoing investigation. Coupling WTS and scRNAseq can reconstruct the immune TME and provide prognostic clinical information, but it does not confirm the functional impact of predicted relationships. Preclinical models to functionalize the study of immune TME manipulation remains a problem in immunomodulatory drug development, including GEA (84–86). We had intentionally structured our trial with serial timepoints to partly address this limitation and enhance confidence in our observations and we hope the future work with improved model systems can test some of our observations (87). We note that the focus of our work is for hypothesis generation for future studies and future work may expand on the number of patients analyzed. We provide in depth analyses of primary lesions but acknowledge the TME of metastatic lesions may be different and further work is needed to characterize metastatic sites in treatment response. In addition, while we present analyses on the immune interaction networks, detailed analyses incorporating tumor genetics, neoantigen prediction, and T-cell receptor sequences are beyond the scope of this manuscript but remain areas of interest. In fact, overall, we envision our dataset to serve as an in silico framework to directly support and evaluate observations from investigators exploring preclinical models in GEA and hope our data will be a valuable resource for the field.
METHODS
Clinical Trial
All patients were enrolled in this prospective open-label phase II trial (ClinicalTrial.gov identifier: NCT04249739). Eligible patients were required to meet the following criteria: (i) at least 19 years old, (ii) histologically confirmed diagnosis of unresectable, metastatic gastric cancer, (iii) adequate organ function per protocol, and (iv) Eastern Cooperative Oncology Group performance status of 0 or 1 (Supplementary Table S8). All patients were naïve to prior chemotherapy. The trial protocol was approved by the Institutional Review Board (IRB) of Samsung Medical Center (Seoul, Korea; IRB No. 2019–11–089) and was conducted in accordance with the Declaration of Helsinki and the Guidelines for Good Clinical Practice. All patients provided written informed consent before enrollment. If the tumor was HER2−, the patient was enrolled on to capecitabine/oxaliplatin/pembrolizumab. If the tumor was HER2+, the patient received capecitabine/cisplatin/trastuzumab and pembrolizumab. The trial was registered at clinicaltrials.gov and the full protocol is provided in the Supplementary Material.
Tumor Sample Collection
All primary tissues were collected from the patients who had biopsies at day 1 before cycle 1, day 1 before cycle 2 and day 1 before cycle 7. Tumor tissues were obtained from endoscopic site-mapping biopsies. If tumor purity was estimated to be more than 40% after pathologic reviews, tumor DNA and RNA were extracted by using a QIAamp Mini Kit (Qiagen) according to the manufacturers’ instructions for exome and transcriptome sequencing. Concentration, 260/280 and 260/230nm ratios were measured with ND1000 spectrophotometer (Nanodrop Technologies, Thermo Fisher Scientific) and then DNA/RNA was quantified using a Qubit fluorometer (Life Technologies).
PD-L1 IHC
Tissues were freshly cut into 4-μm sections, mounted on Fisherbrand Superfrost Plus Microscope Slides (Thermo Fisher Scientific), and then dried at 60°C for 1 hour. IHC staining was carried out on a Dako Autostainer Link 48 system (Agilent Technologies) using the Dako PD-L1 IHC 22C3 pharmDx Kit (Agilent Technologies) with the EnVision FLEX Visualization System and counterstained with hematoxylin according to the manufacturer's instructions. PD-L1 protein expression was determined using CPS, which was the number of PD-L1–stained cells (tumor cells, lymphocytes, and macrophages) divided by the total number of viable tumor cells and multiplied by 100. The specimen was considered to have PD-L1 expression if CPS ≥ 1.
MSI Status
Tumor tissue MSI status was determined using PCR analysis of five markers with mononucleotide repeats (BAT-25, BAT-26, NR-21, NR-24, and NR-27). Briefly, each sense primer was end-labeled with FAM, HEX, or NED. Pentaplex PCR was performed, and the PCR products were run on an Applied Biosystems PRISM 3130 automated genetic analyzer. Allelic sizes were estimated using Genescan 2.1 software (Applied Biosystems). Samples with allelic size variations in more than two microsatellites were considered MSI-H.
Whole Exome and Whole Transcriptome Sequencing
Sequencing was performed using genomic DNA (gDNA) from the tumor tissues and matched blood samples using a QIAamp DNA Blood kit (QIAGEN). For construction of standard exome capture libraries, we used the Agilent SureSelect Target Enrichment protocol for an Illumina paired-end sequencing library together with 1 μg of inputted gDNA. In all samples, the SureSelect Human All Exon V6 probe set was used. We assessed the quantity and quality of DNA by PicoGreen and agarose gel electrophoresis. We diluted 1 μg of gDNA in the elution buffer and sheared it to a target peak size of 150 to 200 bp using the Covaris LE220 focused ultrasonicator device (Covaris Inc.) according to the manufacturer's recommendations. The fragmented DNA was repaired, and “A” was ligated to the 3′-end. Then, we ligated the fragments with Agilent adapters and amplified them using PCR. The prepared libraries were quantified using the TapeStation DNA ScreenTape D1000 (Agilent). For exome capture, 250 ng of DNA library was mixed with hybridization buffer, blocking mixes, RNase block, and 5 μg of SureSelect all exon capture library, according to the standard Agilent SureSelect Target Enrichment protocol. Hybridization to the capture baits was conducted at 65°C using a heated thermal cycler lid option at 105°C for 24 hours on the PCR machine. The captured DNA was washed and amplified. The final purified product was quantified by qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using the TapeStation DNA ScreenTape D1000 (Agilent). Samples were multiplexed, and flow-cell clusters were created using the TruSeq Rapid Cluster kit and the TruSeq Rapid SBS kit (Illumina). Indexed libraries were submitted to an Illumina HiSeq2500 (Illumina), and paired-end (2 × 100 bp) sequencing was performed.
scRNAseq
For single-cell preparation, tumor tissue was dissociated with the gentleMACS Dissociator and Tumor infiltrating Lymphocyte Kit (Miltenyi Biotec) according to the manufacturer's protocol. The cells were then cryopreserved in liquid nitrogen until use. All samples showed a viability of around 90% on average after thawing. We performed 5′ gene expression profiling on the same single-cell suspension using the Chromium Single Cell V(D)J Solution from 10x Genomics according to the manufacturer's instructions. Up to 8,000 cells were loaded onto a 10x Genomics cartridge for each sample. Cell-barcoded 5′ gene expression Libraries were constructed and sequenced at a depth of approximately 50,000 reads per cell using the NovaSeq 6000 platform (Illumina).
WES Analysis
Somatic Variant Calling
WES reads were aligned to the reference human genome GRCh37 using BWA-MEM (88) followed by preprocessing steps, including duplicate marking, indel realignment, and base recalibration using the Genome Analysis Toolkit (GATK; version 4.1.1.0) (89), generating analysis-ready BAM files. To increase the sensitivity for identifying both the lower and higher allele frequencies of somatic variants in the given tumor and paired normal BAM files at the genomic locus, we used the union variant callsets from two tools: MuTect2 (90). Default parameters were applied, and both variant callers were run with dbSNP (version 138; ref. 91), 1000G (phase I; ref. 92), and HapMap (phase III; ref. 93) data for known polymorphic sites. Filtered variants with minimum depth ≥ 5 and minimum alternative alleles ≥ 2 were annotated using the Ensembl Variant Effect Predictor (VEP; release version 87; ref. 94) with the GRCh37 database.
Mutational Signature Analysis
Mutational signature analysis was performed using the deconstructSigs package (version 1.6.0) in R (PMID: 26899170). Exome regions were defined by the Agilent Sureselect V5 target region. Only somatic mutations in exome regions were considered, and trinucleotide counts were normalized by the number of times each trinucleotide context was observed in the exome region. Mutational signatures were represented by the following terms: age (SBS1 and SBS5), apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC; SBS2 and SBS13), UV (SBS7a, SBS7b, SBS7c, and SBS7d), smoking (SBS4), homologous recombination deficiency (HRD; SBS3), mismatch repair deficiency (MMRD; SBS6, SBS15, SBS20, and SBS26), nucleotide excision repair deficiency (NERD; SBS8), DNA proofreading deficiency (DPD; SBS10a and SBS10b), and base excision repair deficiency (BERD; SBS18).
Whole-Transcriptome Sequencing Analysis
We annotated RNA sequence reads with ENSEMBL (version 98) and aligned them to the human reference genome (GRCh38) using STAR (version 2.6.1; ref. 95). We quantified in units of transcript per million (TPM) as a function of gene expression using RSEM (version 1.3.1; ref. 96), applying the option parameters suggested by the GTEx project. TPM values less than one were considered unreliable and substituted with zero. By integrating the transcriptomic data, we classified each tumor sample into two microenvironment subtypes (immune-depleted, immune-enriched) using the molecular functional portrait as previously described (32). Briefly, the samples were classified by TME into 4 categories: depleted, fibrotic, immune-enriched, immune-enriched/fibrotic. We then simplified these classifications into two categories mainly by the immune levels because our research focused on the relation between chemoimmunotherapy and inflammatory remodeling. The GSVA algorithm was used to validate our previous findings in this cohort (97).
scRNA-seq Analysis
Preprocessing and Annotations
We aligned scRNA-seq reads to the GRCh38 human genome reference and quantified them using a Cellranger (version 5.0). The data from all samples were combined in R v.4.1 using the Seurat package v.4.0 (98). We filtered out doublets using Scrublet (99). In addition, cells with low-quality libraries (<400 genes) and high mitochondrial read proportion (>30%) were filtered out. Data from each sample were normalized, scaled, and subjected to principal component analysis, followed by a batch correction using the Harmony (100). We used the Uniform Manifold Approximation and Projection (UMAP) algorithm to reduce the dimension for visual representation, and identified cell clusters using a shared nearest neighbor modularity optimization–based clustering algorithm. We identified various cell type clusters using the “FindAllMarkers” function in Seurat for each cluster and annotated them based on the expression of representative lineage markers. Gene enrichment–related phagocytosis, angiogenesis, and PDGF pathway were estimated using the “AddModuleScore” function in Seurat.
InferCNV
inferCNV (https://github.com/broadinstitute/infercnv) was run on the unprocessed epithelial cells from each scRNA-seq tumor sample individually to infer copy-number variations. When available, a matching normal sample was used as the reference. For the tumor samples without a matching normal sample, a standard reference was generated consisting of a random sampling of 20,000 cells from the combined population of epithelial cells from all normal samples, stratified by patient. The inputs include the combined count matrices of both the sample of interest and reference formatted as an R dgCMatrix, group annotations per cell barcode, and a list of the groups to use as reference. A gene ordering file was generated from the standard hg38 genome reference using the script provided in the above github repo. The i6 HMM implementation of inferCNV was used with a cutoff of 0.1 (as advised for 10x data), the subclusters analysis mode and median filtering enabled. This tool was run on Terra (https://app.terra.bio) via a workflow available here: https://dockstore.org/workflows/github.com/MilanParikh/infercnv_workflow.
Numbat
Numbat (https://github.com/kharchenkolab/numbat/) was run on each full tumor sample (all cell types) individually to call copy-number variations and differentiate normal and tumor cells. When available a matching normal sample was used as the reference. For the tumor samples without a matching normal sample, a standard reference was generated consisting of a random sampling of 25% of all cells from normal samples, about 26,000 cells. The inputs include the bam file, bam index file, 10x barcodes file, and the raw count matrix as an R dgCMatrix for the sample of interest, as well as the cell-type annotations and count matrix as an R dgCMatrix for the reference. All default parameters were used, including the provided SNP VCF and phasing panel from the 1000 Genome project. This tool was run on Terra (https://app.terra.bio) via a workflow available here: https://dockstore.org/workflows/github.com/MilanParikh/numbat_workflow.
Consensus NMF
Consensus NMF (cNMF; https://github.com/dylkot/cNMF) was run on each cell type at each timepoint separately for tumor samples or for each cell type separately in the normal samples to derive gene programs. For each of these runs, the input was an unprocessed AnnData file subset to the population of interest. The custom parameters used include the number of highest variance genes to use in the factorization step (2000) and the range of k values tested for by cNMF (3 to 101 with intervals of 3). All other default parameters were used. Once the prepare, factorization, and combine steps were completed, the generated k selection plot (plotting stability or error vs. k values) was used to manually select the most appropriate k value, and then run the consensus step. To filter overlapping programs between timepoints or normal/malignant samples within the same cell type, a hypergeometric test was performed between the top 100 genes of each pair of programs. The Bonferroni-corrected P value was calculated and program pairs with a P value of less than or equal to 0.05 were counted as overlapping. This tool was run on Terra (https://app.terra.bio) via a workflow available here: https://portal.firecloud.org/?return=terra#methods/mparikh/cnmf_parallel/7. The cNMF programs were subsequently manually annotated by running GSEA on the top 100 weighted genes for each program using the Enrichr implementation in GSEApy (https://gseapy.readthedocs.io/en/latest/introduction.html), by comparing program correlation in the single-cell data with other known program signatures, and by manually examining the top weighted genes in each program.
To highlight the robustness of the approach for identifying gene programs using consensus nonnegative matrix factorization, we have performed 5-fold cross validation and looked to see how frequently overlapping programs are identified. In detail, we split the full dataset into cross validation folds by randomly splitting patients into five roughly 20% subsets. For each fold, we left one of these subsets out so that each fold contains data from roughly 80% of the patients. We further split each fold by cell type (at the level of B cells, epithelial, myeloid, NK cells, stromal, and T cells) and time point/tissue type (baseline tumor, follow-up 1 tumor, follow-up 2 tumor, and all time points normal tissue), as had been done in the cNMF analysis across the entire dataset. Across each of these splits and across all folds we performed cNMF to determine covarying gene programs in each. We also performed this process (splitting into cell type and time point and performing cNMF) on the full dataset. cNMF was performed using the same input hyperparameters in all cases, except for the number of programs found in each split, which was tuned for each run individually based on the stability and error rate of the cNMF solution. For each cell type and tissue type/time point split across all the cross-validation folds (in addition to the combined data set), the top 100 genes were selected for each gene program identified. Then, for each corresponding cell type split across the cross-validation folds, the amount of overlap in the gene lists for the programs in a particular split were measured using a hypergeometric test to determine whether there was a significant overlap between two particular gene program lists belonging to different folds. Programs identified for any time point/tissue type were considered for these comparisons.
Covarying Programs
To find sets of covarying gene programs, or hubs, a patient by program matrix was generated for each timepoint where the values were the 90th percentile of the cNMF program score for that program and patient. This was repeated for the 25th, 50th, 75th, and 95th percentiles as well. For each of these matrices a pairwise correlation of columns was calculated using pandas’ DataFrame.corr() method, resulting in a program-by-program correlation matrix. This was then visualized using Seaborn's clustermap function with Euclidean distance as the spatial distance metric. The 90th percentile data is displayed in Figs. 3 and 6 and was used to identify groups of covarying programs. To find sets of covarying gene programs, or hubs, a patient by program matrix was generated for each timepoint where the values were the 90th percentile of the cNMF program score for that program and patient. The Pearson correlation coefficient, R, was calculated for each pair of programs using pandas’ Dataframe.corr() method. To calculate significance, the patient assignments for each cell were randomly shuffled using pandas’ Series.sample() method and then the 90th percentile scores and correlation coefficients were recalculated. This was repeated for 1,000 iterations to generate a null distribution of correlations for each pair of programs. A p-value was then calculated by counting how many times the randomly shuffled patient assignment correlations were higher than the true correlation, divided by the total number of iterations.
Ligand–receptor Analysis
CellphoneDB's (https://github.com/ventolab/CellphoneDB) statistical analysis method was run on each time point and normal cells separately to infer ligand–receptor pairs. The inputs include an Anndata file (.h5ad) for each timepoint and cell type annotations provided as a two-column csv with cell barcodes and their corresponding cell type. The default parameters were used including 1,000 iterations, a P value of 0.05, and a threshold of 0.01, with no subsetting. This tool was run on Terra (https://app.terra.bio) via a workflow available here: https://dockstore.org/workflows/github.com/MilanParikh/cellphonedb_workflow.
Statistical Analyses
All statistical analyses were conducted using R V.4.1.1 (The R Foundation for Statistical Computing, www.R-project.org) or as specifically specified for each method used. All variables were compared using Wilcoxon signed-rank test and paired Wilcoxon signed-rank test. All P values were two-sided, and results were determined to be significant at P < 0.05.
Multiplex IHC Analysis with 17 Cell Markers
Five-micron–thick tissue sections were made from formalin-fixed, paraffin-embedded tissue blocks. Tissue sections were mounted on glass slides and were prepared according to the manufacturer's instructions. Tissue sections were baked at 60°C for 2 hours and deparaffinized using Neo-Clear (Sigma Aldrich) and rehydrated in descending grades of ethanol (100%, 95%, 80%, 70% of ethanol) for 5 minutes each. Antigen retrieval was performed under 96°C for 30 minutes (IHC-Tek IHC WORLD). After that, tissues were cooled down to 70°C and washed using deionized water. Before staining, tissues were blocked using 3% BSA for 45 minutes at room temperature in the hydration chamber. The tissues were incubated overnight with antibody cocktails including 17 antibodies (Extended Data Table 1) at 4°C in the hydration chamber. After staining, tissues were washed in 0.2% Triton X-100 (Sigma Aldrich) dissolved in PBS two times for 8 minutes. Tissues were also stained in Ir-intercalator solution which was diluted 1:400 in PBS to detect nuclei. Stained tissues were analyzed using the multiplex IHC (IMC), Hyperion (Standard BioTools Inc) after tuning the instrument according to the manufacturer's instructions. 700 μm × 700 μm regions were selected and analyzed for further analysis. IMC image data were acquired at a laser frequency of 50 Hz. Raw image data were preprocessed using MCD Viewer (Standard BioTools Inc).
Cell segmentation was performed using Highplex FL algorithms in HALO v3.4 (Indica Labs) based on nuclei staining (Ir-intercalator solution). After classification, cells were classified into two groups, tumor and stromal, using a random forest model. We also annotated macrophage M1 and M2, CD8 T cells, CD4 T cells based on signals of each immune cell marker. First, tumor cells were annotated based on the signal of pankeratin (panCK) and these cells were excluded for further cell annotation. CD14-positive and CD68-positive cells were annotated to M1 macrophages derived from monocytes and CD68-positive CD163-negative cells were annotated to M1 macrophages. M2 macrophages were annotated with CD163-positive and CD68-positive cells. CD8-positive cells were annotated to CD8 T cells and CD4-positive cells were annotated to CD4 T cells. All cell segmentation was performed using Highplex FL algorithms. After cell segmentation, the number of each cell was acquired from HALO software and multiplex images of each tissue were visualized in HALO software. All statistical analysis was performed using R software.
Data Availability
Sequencing data from the patient cohort are deposited in the European Nucleotide Archive (ENA) under accession PRJEB60680 and these data will be provided upon reasonable written request for academic use and within the limitations of the clinical trial informed consent and general data protection regulations. All other data is provided within the article and Supplementary Tables. Written requests will be reviewed, and data agreements will be needed.
Supplementary Material
Patient demographics
Adverse events on single arm sequential CAPOX and pembrolizumab in advanced gastric cancer.
Detailed cell type proportions
Annotated cNMF programs
Significant covarying programs
CellPhoneDB significant ligand-receptor interactions
CellPhoneDB unique ligand receptor interactions
Representativeness of study participants
(A) Swimmers plot of patient response and duration in the described clinical trial. (B) Scatter plot showing temporal course of tumor response measured by RECIST for all patients in the reported clinical trial. (C) Progression free survival (PFS) and (D) overall survival (OS) for all patients in the reported clinical trial separated by fast and slow progressors.
Figure S2. (A) CoMut plot demonstrating TCGA subtype, tumor mutational burden, mutational signature enrichment and mutational profiles of key driver genes for all patient samples at baseline in the reported clinical trial. (B) PD-L1 expression for each patient sample at baseline in the reported clinical trial.
Figure S3. (A) Enrichment of gene expression signatures for TME subtypes using bulk RNA-sequencing data for all patient samples at baseline and after chemotherapy (follow-up 1; FU1). (B) Pathway enrichment analysis on differentially expressed genes comparing bulk RNA-seq profiles for samples obtained pretreatment (baseline) and samples obtained after 1 cycle of chemotherapy (FU1). (C) Plots demonstrating change in immune related gene signatures (Treg, Treg trafficking, Th2, immune checkpoints, M2 macrophages, pro-tumorgenic cytokines) after chemotherapy treatment. Lines connect samples obtained from the same patient. Statistical comparison performed using a Wilcoxon signed-rank test. (D) Quantification of CD8+ cell abundance using immunohistochemistry (IHC) at baseline and after chemotherapy (FU1) in 12 patient samples. Statistical comparison performed using a Wilcoxon signed-rank test.
Figure S4. Multiplexed immunofluorescence (mIF) images of BL and FU1 samples from two patients, E17 (slow progressor) and E35 (fast progressor), staining for (A) panCK, PD-L1, CD163, CD68, CD14 and CD8, and (B) panCK, PD-L1, CD4, CD8 and Granzyme B.
Figure S5. Proportion of cells expression each of the 16 markers used for multiplex immunohistochemistry (mIHC) split by timepoint and fast vs slow progressor status.
Figure S6. (A) UMAP embedding of single cell transcriptomes obtained from all samples in this trial. Labeled are canonical cell types separated by tumor v.s. normal tissue, and by timepoint. (B) Heatmap and (C) UMAP embeddings showing marker gene expression for broad cell types in (A). (D) Broad cell type composition per sample across tumor and normal tissue. (E) Cell type proportions from scSeq data by timepoint in tumor and normal tissue excluding HER2+ patients. (F) Cell type proportions from scSeq data by timepoint and TCGA classification excluding HER2+ patients.
Figure S7. (A) UMAP embedding of single cell transcriptomes from epithelial cells obtained from all samples in this trial. Shown are UMAP embeddings with expression of key epithelial genes. (B) UMAP embeddings color coded by predicted annotation of tumor cell v.s. normal cell using Numbat.
Figure S8. Copy number profiles of all epithelial cells from each patient’s samples inferred using inferCNV.
Figure S9. (A) Cell type proportions from scSeq shown by individual patient split by individual high-level cell types across timepoints, color coded by fast vs slow progression status. (B) Cell type proportions from scSeq data by timepoint and fast vs slow progressor status including only GS and CIN patients.
Figure S10. Mean usage of epithelial cNMF programs per sample at (A) baseline, (B) post-chemotherapy (FU1) and (C) post-immunotherapy (FU2).
Figure S11. Mean usage of myeloid cNMF programs per myeloid cell subset at (A) baseline, (B) post-chemotherapy (FU1) and (C) post-immunotherapy (FU2). Mean usage of T cell cNMF programs per T cell cell subset at (D) baseline, (E) postchemotherapy (FU1) and (F) post-immunotherapy (FU2).
Figure S12. Mean usage of stromal cNMF programs per stromal cell subset at (A) baseline, (B) post-chemotherapy (FU1) and (C) post-immunotherapy (FU2).
Figure S13. Boxplots showing usage of epithelial cNMF programs in epithelial cells of fast and slow progressors (A) post-chemotherapy (FU1) and (B) postimmunotherapy (FU2).
Figure S14. cNMF was performed on epithelial cells at baseline. Shown is the mean usage of each cNMF gene program in the epithelial cells of fast and slow progressing patients.
Figure S15. Heatmap showing pairwise correlation of gene program activities across all patient samples at baseline using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK and stromal cells. Hierarchical clustering was performed to identify clusters of co-varying proteins.
Figure S16. Heatmap showing pairwise correlation of gene program activities across all patient samples post-chemotherapy (FU1) using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK and stromal cells. Hierarchical clustering was performed to identify clusters of co-varying proteins.
Figure S17. Heatmap showing pairwise correlation of gene program activities across all patient samples post-immunotherapy (FU2) using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK and stromal cells. Hierarchical clustering was performed to identify clusters of co-varying proteins.
Figure S18. (A) UMAP embedding of single cell transcriptomes of all myeloid cells obtained from all samples in this trial. Labeled are granular myeloid cell types separated by tumor v.s. normal tissue, and by timepoint. (B) UMAP embeddings showing marker gene expression for granular myeloid cell types in (A).
Figure S19. (A) UMAP embedding of single cell transcriptomes of all macrophages obtained from all samples in this trial. Labeled are granular macrophage subsets separated by tumor v.s. normal tissue, and by timepoint. (B) UMAP embeddings showing marker gene expression for macrophage subsets in (A). (C) Relative M1 macrophage proportion in fast and slow progressing patients separated by timepoint. (D) Macrophage subtype proportions at baseline and FU1 in fast versus slow progressing patients, and in non-responder and responder patients. (E) Relative M1 macrophage proportion in non-responder and responder patients separated by timepoint. (F) Proportion of SPP1+ macrophages in nonresponder and responder patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (G) Change in relative M1 proportion from BL to FU1 plotted against change in tumor volume after 1 cycle of chemotherapy, segregated by non-responder and responder patients. (H) GSVA performed for M2 signatures on bulk RNA-seq samples split by timepoints and progression status.
Figure S20. (A) CT imaging of tumor burden in a pair of patients classified as either a responder with increased relative M1 ration or non-responder with unchanged relative M1 ration. (B) CT scans of a second pair of patients as in (A). (C) Multiplexed immunofluorescence (mIF) images of BL, FU1 and FU2 samples from two patients, E17 (slow progressor) and E35 (fast progressor), staining for panCK, PD-L1, CD163, CD68, CD14 and CD8.
Figure S21. (A) Change in M2 ratio from baseline to FU1 in patients with decreased or increased stromal proportions. (B) Proportion of stromal cells in patient samples from fast and slow progressing patients split by timepoint. (C) UMAP embedding of single cell transcriptomes of all stromal cells obtained from all samples in this trial. Labeled are granular stromal cell types separated by tumor v.s. normal tissue, and by timepoint. (D) UMAP embeddings showing marker gene expression for granular stromal cell types in (C). (E) Proportion of myofibroblasts in patient samples from fast and slow progressing patients split by timepoint.
Figure S22. Phagocytosis, angiogenesis and PDGF pathway gene module scores in different M2 macrophage subsets from scRNAseq data.
Figure S23. Heatmaps showing the number of significant inferred ligandreceptor interactions between major cell types using CellPhoneDB on scRNAseq data in (A) normal, (B) baseline tumor, (C) post-chemotherapy (FU1) tumor and (D) post-immunotherapy (FU2) tumor samples.
Figure S24. (A) Immune gene expression program signature scored from bulk RNA-seq profiles across all samples separated by timepoint. (B) Immunogenic cell death signatures scored from bulk RNA-seq profiles split by fast and slow progressing patients and by timepoint.
Figure S25. (A) UMAP embedding of single cell transcriptomes of all T cells obtained from all samples in this trial. Labeled are granular T cell subsets separated by tumor v.s. normal tissue, and by timepoint. (B) UMAP embeddings showing marker gene expression for T cell subsets in (A).
Figure S26. (A) Proportion of CD8 T cells in non-responder and responder patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (B) Multiplexed immunofluorescence (mIF) images of BL, FU1 and FU2 samples from two patients, E40 (slow progressor) and E41 (fast progressor), staining for panCK, PD-L1, CD4, CD8 and Granzyme B. (C) Proportion of CD8 T cells macrophages, obtained from mIF images, at BL, FU1 and FU2 in non-responder and responder patients. Statistical comparison performed using a Wilcoxon signed-rank test. (D) Proportion of naive, memory, effector and exhausted CD8 T cell subsets in non-responder and responder patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (E) Proportion of regulatory T cell and Th1 CD4 T cell subsets in fast v.s. slow progressor patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (F) Expression of CXCL13 in CD4 T cells in fast v.s. slow progressor patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test.
Figure S27. Boxplots showing usage of T cell cNMF programs in T cells of fast and slow progressors (A) post-chemotherapy (FU1) and (B) post-immunotherapy (FU2). (C) Expression of CD39, CD103, CXCL13, 4-1BB and GITR in single CD8 T cells segregated by timepoint (BL, FU1, FU2) and by fast versus slow progressing patients. Statistical comparison performed using a Wilcoxon signedrank test.
Acknowledgments
This work was supported by Dana Farber Cancer Institute/Harvard CancerCare GI SPORE Career Enhancement Award (to A. Mehta), Sky Foundation Pancreatic Cancer Research Grant (to A. Mehta), Doris Duke Charitable Foundation Physician Scientist Fellowship (to A. Mehta), the DeGregorio Family Foundation (to S.J. Klempner), The Torrey Coast Foundation (to S.J. Klempner), Stand Up to Cancer Gastric Cancer Interception Research Team Grant (SU2C-AACR-DT-30–20; to H. Lee, S.J. Klempner, J. Lee). This research was supported by the Bio&Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (grant number: RS-2023-00222838 to S. Kim) and SKKU Excellence in Research Award Research Fund, Sungkyungkwan University, 2022 (to J. Lee). Supported in part by a research grant from Investigator-Initiated Studies Program of MSD (to J. Lee)
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
A. Mehta reports consultant/advisory role for Third Rock Ventures, Asher Biotherapeutics, Abata Therapeutics, ManaT Bio, Flare Therapeutics, venBio Partners, BioNTech, Rheos Medicines and Checkmate Pharmaceuticals. A. Mehta is also currently a part-time Entrepreneur in Residence at Third Rock Ventures, is an equity holder in ManaT Bio, Asher Biotherapeutics and Abata Therapeutics, and received research funding support from Bristol-Myers Squibb. M. Parikh reports personal fees from Third Rock Ventures outside the submitted work. W. Park reports personal fees from Geninus Inc. outside the submitted work. S.J. Klempner reports non-financial support from Arcus and Leap Therapeutics; personal fees from Amgen, AstraZeneca, Sanofi-Aventis, I-Mab Therapeutics, Merck, BMS, Research To Practice, Astellas, Daiichi-Sankyo; other support from Nuvalent, and other support from Turning Point outside the submitted work. J. Lee reports non-financial support from Revolution Medicine, Immodulon, Mirati Therapeutics, and Trutino Biosciences; grants from Astra Zeneca, and grants from Merck Sharp & Dohme outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
M. An: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Mehta: Data curation, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. B. Min: Resources, investigation, writing–review and editing. Y. Heo: Formal analysis, investigation, visualization, methodology, writing–review and editing. S.J. Wright: Visualization, methodology, writing–review and editing. M. Parikh: Visualization, methodology, writing–review and editing. L. Bi: Visualization, methodology, writing–review and editing. H. Lee: Resources, investigation, writing–review and editing. T. Kim: Resources, investigation, writing–review and editing. S. Lee: Resources, formal analysis, investigation, writing–review and editing. J. Moon: Data curation, formal analysis, writing–review and editing. R.J. Park: Formal analysis, visualization, writing–review and editing. M.R. Strickland: Formal analysis, writing–review and editing. W. Park: Data curation, formal analysis, methodology, writing–review and editing. W. Kang: Resources, investigation, writing–review and editing. K. Kim: Resources, data curation, formal analysis, supervision, validation, methodology, writing–review and editing. S. Kim: Conceptualization, resources, formal analysis, supervision, investigation, writing–original draft, writing–review and editing. S.J. Klempner: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing. J. Lee: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing.
References
- 1. Yoon HH, Jin Z, Kour O, Kankeu Fonkoua LA, Shitara K, Gibson MK, et al. Association of PD-L1 expression and other variables with benefit from immune checkpoint inhibition in advanced gastroesophageal cancer: systematic review and meta-analysis of 17 phase 3 randomized clinical trials. JAMA Oncol 2022;8:1456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shitara K, Ajani JA, Moehler M, Garrido M, Gallardo C, Shen L, et al. Nivolumab plus chemotherapy or ipilimumab in gastro-oesophageal cancer. Nature 2022;603:942–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim R, An M, Lee H, Mehta A, Heo YJ, Kim K-M, et al. Early tumor-immune microenvironmental remodeling and response to first-line fluoropyrimidine and platinum chemotherapy in advanced gastric cancer. Cancer Discov 2022;12:984–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang X, Li M, Wu X, Guo T, Zhang L, Tang L, et al. Neoadjuvant PD-1 blockade plus chemotherapy induces a high pathological complete response rate and anti-tumor immune subsets in clinical stage III gastric cancer. Oncoimmunology 2022;11:2135819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xing X, Shi J, Jia Y, Dou Y, Li Z, Dong B, et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in gastric cancer as determined by multiplex immunofluorescence and T cell receptor repertoire analysis. J Immunother Cancer 2022;10:e003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elhanani O, Ben-Uri R, Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 2023;41:404–20. [DOI] [PubMed] [Google Scholar]
- 7. Zaitsev A, Chelushkin M, Dyikanov D, Cheremushkin I, Shpak B, Nomie K, et al. Precise reconstruction of the TME using bulk RNA-seq and a machine learning algorithm trained on artificial transcriptomes. Cancer Cell 2022;40:879–94. [DOI] [PubMed] [Google Scholar]
- 8. Li Z, Gao X, Peng X, May Chen MJ, Li Z, Wei B, et al. Multi-omics characterization of molecular features of gastric cancer correlated with response to neoadjuvant chemotherapy. Sci Adv 2020;6:eaay4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schürch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 2020;183:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V, et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 2021;184:4734–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hwang WL, Jagadeesh KA, Guo JA, Hoffman HI, Yadollahpour P, Reeves JW, et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat Genet 2022;54:1178–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nirmal AJ, Maliga Z, Vallius T, Quattrochi B, Chen AA, Jacobson CA, et al. The spatial landscape of progression and immunoediting in primary melanoma at single-cell resolution. Cancer Discov 2022;12:1518–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 2019;8:e43803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang Y-K, Chen L-T, Ryu M-H, Oh D-Y, Oh SC, Chung HC, et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): a randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2022;23:234–47. [DOI] [PubMed] [Google Scholar]
- 15. Network CGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 17. Kim ST, Cristescu R, Bass AJ, Kim K-M, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. [DOI] [PubMed] [Google Scholar]
- 18. Kwon M, An M, Klempner SJ, Lee H, Kim K-M, Sa JK, et al. Determinants of response and intrinsic resistance to PD-1 blockade in microsatellite instability-high gastric cancer. Cancer Discov 2021;11:2168–85. [DOI] [PubMed] [Google Scholar]
- 19. Luo J, Wu S, Rizvi H, Zhang Q, Egger JV, Osorio JC, et al. Deciphering radiological stable disease to immune checkpoint inhibitors. Ann Oncol 2022;33:824–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun J-M, Shen L, Shah MA, Enzinger P, Adenis A, Doi T, et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study. Lancet 2021;398:759–71. [DOI] [PubMed] [Google Scholar]
- 22. Tian J, Chen JH, Chao SX, Pelka K, Giannakis M, Hess J, et al. Combined PD-1, BRAF and MEK inhibition in BRAFV600E colorectal cancer: a phase 2 trial. Nat Med 2023;29:458–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Topp B, Snyder A, Wolchok J. RECISTv1.1 progression in oncology: Shades of gray. Cancer Cell 2023;41:1003–5. [DOI] [PubMed] [Google Scholar]
- 24. O'Rourke CJ, Salati M, Rae C, Carpino G, Leslie H, Pea A, et al. Molecular portraits of patients with intrahepatic cholangiocarcinoma who diverge as rapid progressors or long survivors on chemotherapy. Gut 2023Sep 27 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janjigian YY, Sanchez-Vega F, Jonsson P, Chatila WK, Hechtman JF, Ku GY, et al. Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov 2018;8:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun K, Xu R, Ma F, Yang N, Li Y, Sun X, et al. scRNA-seq of gastric tumor shows complex intercellular interaction with an alternative T cell exhaustion trajectory. Nat Commun 2022;13:4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M, et al. Single-cell Atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov 2022;12:670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang M, Hu S, Min M, Ni Y, Lu Z, Sun X, et al. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut 2021;70:464–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao T, Soldatov R, Sarkar H, Kurkiewicz A, Biederstedt E, Loh P-R, et al. Haplotype-aware analysis of somatic copy number variations from single-cell transcriptomes. Nat Biotechnol 2023;41:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Combes AJ, Samad B, Krummel MF. Defining and using immune archetypes to classify and treat cancer. Nat Rev Cancer 2023;23:491–505. [DOI] [PubMed] [Google Scholar]
- 32. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021;39:845–65. [DOI] [PubMed] [Google Scholar]
- 33. Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barkley D, Moncada R, Pour M, Liberman DA, Dryg I, Werba G, et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat Genet 2022;54:1192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leung WK, Yu J, Chan FKL, To KF, Chan MWY, Ebert MPA, et al. Expression of trefoil peptides (TFF1, TFF2, and TFF3) in gastric carcinomas, intestinal metaplasia, and non-neoplastic gastric tissues. J Pathol 2002;197:582–8. [DOI] [PubMed] [Google Scholar]
- 37. Song J-Y, Kim B-W, Lee A-W, Lee K-Y, Chung I-S, Lee B-I, et al. Expression of MUC5AC and trefoil peptide 1 (TFF1) in the subtypes of intestinal metaplasia. Clin Endosc 2012;45:151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lenos KJ, Bach S, Ferreira Moreno L, Ten Hoorn S, Sluiter NR, Bootsma S, et al. Molecular characterization of colorectal cancer related peritoneal metastatic disease. Nat Commun 2022;13:4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsu H-J, Tung C-P, Yu C-M, Chen C-Y, Chen H-S, Huang Y-C, et al. Eradicating mesothelin-positive human gastric and pancreatic tumors in xenograft models with optimized anti-mesothelin antibody–drug conjugates from synthetic antibody libraries. Sci Rep 2021;11:15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Einama T, Homma S, Kamachi H, Kawamata F, Takahashi K, Takahashi N, et al. Luminal membrane expression of mesothelin is a prominent poor prognostic factor for gastric cancer. Br J Cancer 2012;107:137–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanada K-I, Zhao C, Gil-Hoyos R, Gartner JJ, Chow-Parmer C, Lowery FJ, et al. A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers. Cancer Cell 2022;40:479–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu B, Zhang Y, Wang D, Hu X, Zhang Z. Single-cell meta-analyses reveal responses of tumor-reactive CXCL13 T cells to immune-checkpoint blockade. Nat Cancer 2022;3:1123–36. [DOI] [PubMed] [Google Scholar]
- 43. Kersten K, Hu KH, Combes AJ, Samad B, Harwin T, Ray A, et al. Spatiotemporal co-dependency between macrophages and exhausted CD8+ T cells in cancer. Cancer Cell 2022;40:624–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov 2022;21:799–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nixon BG, Kuo F, Ji L, Liu M, Capistrano K, Do M, et al. Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity 2022;55:2044–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021;184:792–809. [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y, Chen H, Mo H, Hu X, Gao R, Zhao Y, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 2021;39:1578–93. [DOI] [PubMed] [Google Scholar]
- 49. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB v2.0: Inferring cell-cell communication from combined expression of multi-subunit receptor-ligand complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 50. Yang R, Sun L, Li CF, Wang YH, Yao J, Li H. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun 2021;12:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ott PA, Bang Y-J, Piha-Paul SA, Razak ARA, Bennouna J, Soria J-C, et al. T-Cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 Cancers: KEYNOTE-028. J Clin Oncol 2019;37:318–27. [DOI] [PubMed] [Google Scholar]
- 53. Cristescu R, Nebozhyn M, Zhang C, Albright A, Kobie J, Huang L, et al. Transcriptomic determinants of response to pembrolizumab monotherapy across solid tumor types. Clin Cancer Res 2022;28:1680–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97–111. [DOI] [PubMed] [Google Scholar]
- 55. Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huseni MA, Wang L, Klementowicz JE, Yuen K, Breart B, Orr C, et al. CD8+ T cell-intrinsic IL-6 signaling promotes resistance to anti-PD-L1 immunotherapy. Cell Rep Med 2022;4:100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fisher DT, Appenheimer MM, Evans SS. The two faces of IL-6 in the tumor microenvironment. Semin Immunol 2014;26:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Revel M, Sautès-Fridman C, Fridman W-H, Roumenina LT. C1q+ macrophages: passengers or drivers of cancer progression. Trends Cancer Res 2022;8:517–26. [DOI] [PubMed] [Google Scholar]
- 59. Xiao G, Deng A, Liu H, Ge G, Liu X. Activator protein 1 suppresses antitumor T-cell function via the induction of programmed death 1. Proc Natl Acad Sci U S A 2012;109:15419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mullins RDZ, Pal A, Barrett TF, Heft Neal ME, Puram SV. Epithelial-mesenchymal plasticity in tumor immune evasion. Cancer Res 2022;82:2329–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang M, Chen X, Tan P, Wang Y, Pan X, Lin T, et al. Acquired semi-squamatization during chemotherapy suggests differentiation as a therapeutic strategy for bladder cancer. Cancer Cell 2022;40:1044–59. [DOI] [PubMed] [Google Scholar]
- 62. Wolf DM, Yau C, Wulfkuhle J, Brown-Swigart L, Gallagher RI, Lee PRE, et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: Presssdictive biomarkers across 10 cancer therapies. Cancer Cell 2022;40:609–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li K, Tandurella JA, Gai J, Zhu Q, Lim SJ, Thomas DL II, et al. Multi-omic analyses of changes in the tumor microenvironment of pancreatic adenocarcinoma following neoadjuvant treatment with anti–PD-1 therapy. Cancer Cell 2022;40:1374–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen B, Scurrah CR, McKinley ET, Simmons AJ, Ramirez-Solano MA, Zhu X, et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 2021;184:6262–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hingorani SR. Epithelial and stromal co-evolution and complicity in pancreatic cancer. Nat Rev Cancer 2023;23:57–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Labrie M, Brugge JS, Mills GB, Zervantonakis IK. Therapy resistance: opportunities created by adaptive responses to targeted therapies in cancer. Nat Rev Cancer 2022;22:323–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim TK, Vandsemb EN, Herbst RS, Chen L. Adaptive immune resistance at the tumour site: mechanisms and therapeutic opportunities. Nat Rev Drug Discov 2022;21:529–40. [DOI] [PubMed] [Google Scholar]
- 68. Shi T, Zhang Y, Wang Y, Song X, Wang H, Zhou X, et al. DKK1 promotes tumor immune evasion and impedes anti–PD-1 treatment by inducing immunosuppressive macrophages in gastric cancer. Cancer Immunol Res 2022;10:1506–24. [DOI] [PubMed] [Google Scholar]
- 69. Hong C, Schubert M, Tijhuis AE, Requesens M, Roorda M, van den Brink A, et al. cGAS–STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 2022;607:366–73. [DOI] [PubMed] [Google Scholar]
- 70. Wang S, Campos J, Gallotta M, Gong M, Crain C, Naik E, et al. Intratumoral injection of a CpG oligonucleotide reverts resistance to PD-1 blockade by expanding multifunctional CD8+ T cells. Proc Natl Acad Sci U S A 2016;113:E7240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sato-Kaneko F, Yao S, Ahmadi A, Zhang SS, Hosoya T, Kaneda MM, et al. Combination immunotherapy with TLR agonists and checkpoint inhibitors suppresses head and neck cancer. JCI Insight 2017;2:e93397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Veneziani I, Alicata C, Pelosi A, Landolina N, Ricci B, D'Oria V, et al. Toll-like receptor 8 agonists improve NK-cell function primarily targeting CD56brightCD16- subset. J Immunother Cancer 2022;10:e003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cytryn SL, Moy RH, Cowzer D, Shah RH, Chou JF, Joshi SS, et al. First-line regorafenib with nivolumab and chemotherapy in advanced oesophageal, gastric, or gastro-oesophageal junction cancer in the USA: a single-arm, single-centre, phase 2 trial. Lancet Oncol 2023;24:1073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dias Costa A, Väyrynen SA, Chawla A, Zhang J, Väyrynen JP, Lau MC, et al. Neoadjuvant chemotherapy is associated with altered immune cell infiltration and an anti-tumorigenic microenvironment in resected pancreatic cancer. Clin Cancer Res 2022;28:5167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Grout JA, Sirven P, Leader AM, Maskey S, Hector E, Puisieux I, et al. Spatial positioning and matrix programs of cancer-associated fibroblasts promote T-cell exclusion in human lung tumors. Cancer Discov 2022;12:2606–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bortolomeazzi M, Keddar MR, Montorsi L, Acha-Sagredo A, Benedetti L, Temelkovski D, et al. Immunogenomics of colorectal cancer response to checkpoint blockade: analysis of the KEYNOTE 177 Trial and validation cohorts. Gastroenterology 2021;161:1179–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11:889–96. [DOI] [PubMed] [Google Scholar]
- 78. Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CMT, Pryer N, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 2014;26:623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Peranzoni E, Lemoine J, Vimeux L, Feuillet V, Barrin S, Kantari-Mimoun C, et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti–PD-1 treatment. Proc Natl Acad Sci U S A 2018;115:E4041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fan Y, Li Y, Yao X, Jin J, Scott A, Liu B, et al. Epithelial SOX9 drives progression and metastases of gastric adenocarcinoma by promoting immunosuppressive tumour microenvironment. Gut 2023;72:624–37. [DOI] [PubMed] [Google Scholar]
- 81. Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 2020;181:442–59. [DOI] [PubMed] [Google Scholar]
- 82. Liu Y, Zhang Q, Xing B, Luo N, Gao R, Yu K, et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell 2022;40:424–37. [DOI] [PubMed] [Google Scholar]
- 83. Yaddanapudi K, Putty K, Rendon BE, Lamont GJ, Faughn JD, Satoskar A, et al. Control of tumor-associated macrophage alternative activation by macrophage migration inhibitory factor. J Immunol 2013;190:2984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ho WW, Pittet MJ, Fukumura D, Jain RK. The local microenvironment matters in preclinical basic and translational studies of cancer immunology and immunotherapy. Cancer Cell 2022;40:701–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Reardon DA, Gokhale PC, Klein SR, Ligon KL, Rodig SJ, Ramkissoon SH, et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol Res 2016;4:124–35. [DOI] [PubMed] [Google Scholar]
- 86. Horton BL, Morgan DM, Momin N, Zagorulya M, Torres-Mejia E, Bhandarkar V, et al. Lack of CD8+ T cell effector differentiation during priming mediates checkpoint blockade resistance in non-small cell lung cancer. Sci Immunol 2021;6:eabi8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Leibold J, Amor C, Tsanov KM, Ho Y-J, Sánchez-Rivera FJ, Feucht J, et al. Somatic mouse models of gastric cancer reveal genotype-specific features of metastatic disease. Nat Cancer 2024 Jan 4 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. International HapMap 3 Consortium; Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010;467:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient demographics
Adverse events on single arm sequential CAPOX and pembrolizumab in advanced gastric cancer.
Detailed cell type proportions
Annotated cNMF programs
Significant covarying programs
CellPhoneDB significant ligand-receptor interactions
CellPhoneDB unique ligand receptor interactions
Representativeness of study participants
(A) Swimmers plot of patient response and duration in the described clinical trial. (B) Scatter plot showing temporal course of tumor response measured by RECIST for all patients in the reported clinical trial. (C) Progression free survival (PFS) and (D) overall survival (OS) for all patients in the reported clinical trial separated by fast and slow progressors.
Figure S2. (A) CoMut plot demonstrating TCGA subtype, tumor mutational burden, mutational signature enrichment and mutational profiles of key driver genes for all patient samples at baseline in the reported clinical trial. (B) PD-L1 expression for each patient sample at baseline in the reported clinical trial.
Figure S3. (A) Enrichment of gene expression signatures for TME subtypes using bulk RNA-sequencing data for all patient samples at baseline and after chemotherapy (follow-up 1; FU1). (B) Pathway enrichment analysis on differentially expressed genes comparing bulk RNA-seq profiles for samples obtained pretreatment (baseline) and samples obtained after 1 cycle of chemotherapy (FU1). (C) Plots demonstrating change in immune related gene signatures (Treg, Treg trafficking, Th2, immune checkpoints, M2 macrophages, pro-tumorgenic cytokines) after chemotherapy treatment. Lines connect samples obtained from the same patient. Statistical comparison performed using a Wilcoxon signed-rank test. (D) Quantification of CD8+ cell abundance using immunohistochemistry (IHC) at baseline and after chemotherapy (FU1) in 12 patient samples. Statistical comparison performed using a Wilcoxon signed-rank test.
Figure S4. Multiplexed immunofluorescence (mIF) images of BL and FU1 samples from two patients, E17 (slow progressor) and E35 (fast progressor), staining for (A) panCK, PD-L1, CD163, CD68, CD14 and CD8, and (B) panCK, PD-L1, CD4, CD8 and Granzyme B.
Figure S5. Proportion of cells expression each of the 16 markers used for multiplex immunohistochemistry (mIHC) split by timepoint and fast vs slow progressor status.
Figure S6. (A) UMAP embedding of single cell transcriptomes obtained from all samples in this trial. Labeled are canonical cell types separated by tumor v.s. normal tissue, and by timepoint. (B) Heatmap and (C) UMAP embeddings showing marker gene expression for broad cell types in (A). (D) Broad cell type composition per sample across tumor and normal tissue. (E) Cell type proportions from scSeq data by timepoint in tumor and normal tissue excluding HER2+ patients. (F) Cell type proportions from scSeq data by timepoint and TCGA classification excluding HER2+ patients.
Figure S7. (A) UMAP embedding of single cell transcriptomes from epithelial cells obtained from all samples in this trial. Shown are UMAP embeddings with expression of key epithelial genes. (B) UMAP embeddings color coded by predicted annotation of tumor cell v.s. normal cell using Numbat.
Figure S8. Copy number profiles of all epithelial cells from each patient’s samples inferred using inferCNV.
Figure S9. (A) Cell type proportions from scSeq shown by individual patient split by individual high-level cell types across timepoints, color coded by fast vs slow progression status. (B) Cell type proportions from scSeq data by timepoint and fast vs slow progressor status including only GS and CIN patients.
Figure S10. Mean usage of epithelial cNMF programs per sample at (A) baseline, (B) post-chemotherapy (FU1) and (C) post-immunotherapy (FU2).
Figure S11. Mean usage of myeloid cNMF programs per myeloid cell subset at (A) baseline, (B) post-chemotherapy (FU1) and (C) post-immunotherapy (FU2). Mean usage of T cell cNMF programs per T cell cell subset at (D) baseline, (E) postchemotherapy (FU1) and (F) post-immunotherapy (FU2).
Figure S12. Mean usage of stromal cNMF programs per stromal cell subset at (A) baseline, (B) post-chemotherapy (FU1) and (C) post-immunotherapy (FU2).
Figure S13. Boxplots showing usage of epithelial cNMF programs in epithelial cells of fast and slow progressors (A) post-chemotherapy (FU1) and (B) postimmunotherapy (FU2).
Figure S14. cNMF was performed on epithelial cells at baseline. Shown is the mean usage of each cNMF gene program in the epithelial cells of fast and slow progressing patients.
Figure S15. Heatmap showing pairwise correlation of gene program activities across all patient samples at baseline using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK and stromal cells. Hierarchical clustering was performed to identify clusters of co-varying proteins.
Figure S16. Heatmap showing pairwise correlation of gene program activities across all patient samples post-chemotherapy (FU1) using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK and stromal cells. Hierarchical clustering was performed to identify clusters of co-varying proteins.
Figure S17. Heatmap showing pairwise correlation of gene program activities across all patient samples post-immunotherapy (FU2) using the 90th percentile of patient-level program activity in epithelial, myeloid, T, NK and stromal cells. Hierarchical clustering was performed to identify clusters of co-varying proteins.
Figure S18. (A) UMAP embedding of single cell transcriptomes of all myeloid cells obtained from all samples in this trial. Labeled are granular myeloid cell types separated by tumor v.s. normal tissue, and by timepoint. (B) UMAP embeddings showing marker gene expression for granular myeloid cell types in (A).
Figure S19. (A) UMAP embedding of single cell transcriptomes of all macrophages obtained from all samples in this trial. Labeled are granular macrophage subsets separated by tumor v.s. normal tissue, and by timepoint. (B) UMAP embeddings showing marker gene expression for macrophage subsets in (A). (C) Relative M1 macrophage proportion in fast and slow progressing patients separated by timepoint. (D) Macrophage subtype proportions at baseline and FU1 in fast versus slow progressing patients, and in non-responder and responder patients. (E) Relative M1 macrophage proportion in non-responder and responder patients separated by timepoint. (F) Proportion of SPP1+ macrophages in nonresponder and responder patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (G) Change in relative M1 proportion from BL to FU1 plotted against change in tumor volume after 1 cycle of chemotherapy, segregated by non-responder and responder patients. (H) GSVA performed for M2 signatures on bulk RNA-seq samples split by timepoints and progression status.
Figure S20. (A) CT imaging of tumor burden in a pair of patients classified as either a responder with increased relative M1 ration or non-responder with unchanged relative M1 ration. (B) CT scans of a second pair of patients as in (A). (C) Multiplexed immunofluorescence (mIF) images of BL, FU1 and FU2 samples from two patients, E17 (slow progressor) and E35 (fast progressor), staining for panCK, PD-L1, CD163, CD68, CD14 and CD8.
Figure S21. (A) Change in M2 ratio from baseline to FU1 in patients with decreased or increased stromal proportions. (B) Proportion of stromal cells in patient samples from fast and slow progressing patients split by timepoint. (C) UMAP embedding of single cell transcriptomes of all stromal cells obtained from all samples in this trial. Labeled are granular stromal cell types separated by tumor v.s. normal tissue, and by timepoint. (D) UMAP embeddings showing marker gene expression for granular stromal cell types in (C). (E) Proportion of myofibroblasts in patient samples from fast and slow progressing patients split by timepoint.
Figure S22. Phagocytosis, angiogenesis and PDGF pathway gene module scores in different M2 macrophage subsets from scRNAseq data.
Figure S23. Heatmaps showing the number of significant inferred ligandreceptor interactions between major cell types using CellPhoneDB on scRNAseq data in (A) normal, (B) baseline tumor, (C) post-chemotherapy (FU1) tumor and (D) post-immunotherapy (FU2) tumor samples.
Figure S24. (A) Immune gene expression program signature scored from bulk RNA-seq profiles across all samples separated by timepoint. (B) Immunogenic cell death signatures scored from bulk RNA-seq profiles split by fast and slow progressing patients and by timepoint.
Figure S25. (A) UMAP embedding of single cell transcriptomes of all T cells obtained from all samples in this trial. Labeled are granular T cell subsets separated by tumor v.s. normal tissue, and by timepoint. (B) UMAP embeddings showing marker gene expression for T cell subsets in (A).
Figure S26. (A) Proportion of CD8 T cells in non-responder and responder patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (B) Multiplexed immunofluorescence (mIF) images of BL, FU1 and FU2 samples from two patients, E40 (slow progressor) and E41 (fast progressor), staining for panCK, PD-L1, CD4, CD8 and Granzyme B. (C) Proportion of CD8 T cells macrophages, obtained from mIF images, at BL, FU1 and FU2 in non-responder and responder patients. Statistical comparison performed using a Wilcoxon signed-rank test. (D) Proportion of naive, memory, effector and exhausted CD8 T cell subsets in non-responder and responder patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (E) Proportion of regulatory T cell and Th1 CD4 T cell subsets in fast v.s. slow progressor patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test. (F) Expression of CXCL13 in CD4 T cells in fast v.s. slow progressor patients split by timepoint. Statistical comparisons performed using a Wilcoxon signed-rank test.
Figure S27. Boxplots showing usage of T cell cNMF programs in T cells of fast and slow progressors (A) post-chemotherapy (FU1) and (B) post-immunotherapy (FU2). (C) Expression of CD39, CD103, CXCL13, 4-1BB and GITR in single CD8 T cells segregated by timepoint (BL, FU1, FU2) and by fast versus slow progressing patients. Statistical comparison performed using a Wilcoxon signedrank test.
Data Availability Statement
Sequencing data from the patient cohort are deposited in the European Nucleotide Archive (ENA) under accession PRJEB60680 and these data will be provided upon reasonable written request for academic use and within the limitations of the clinical trial informed consent and general data protection regulations. All other data is provided within the article and Supplementary Tables. Written requests will be reviewed, and data agreements will be needed.