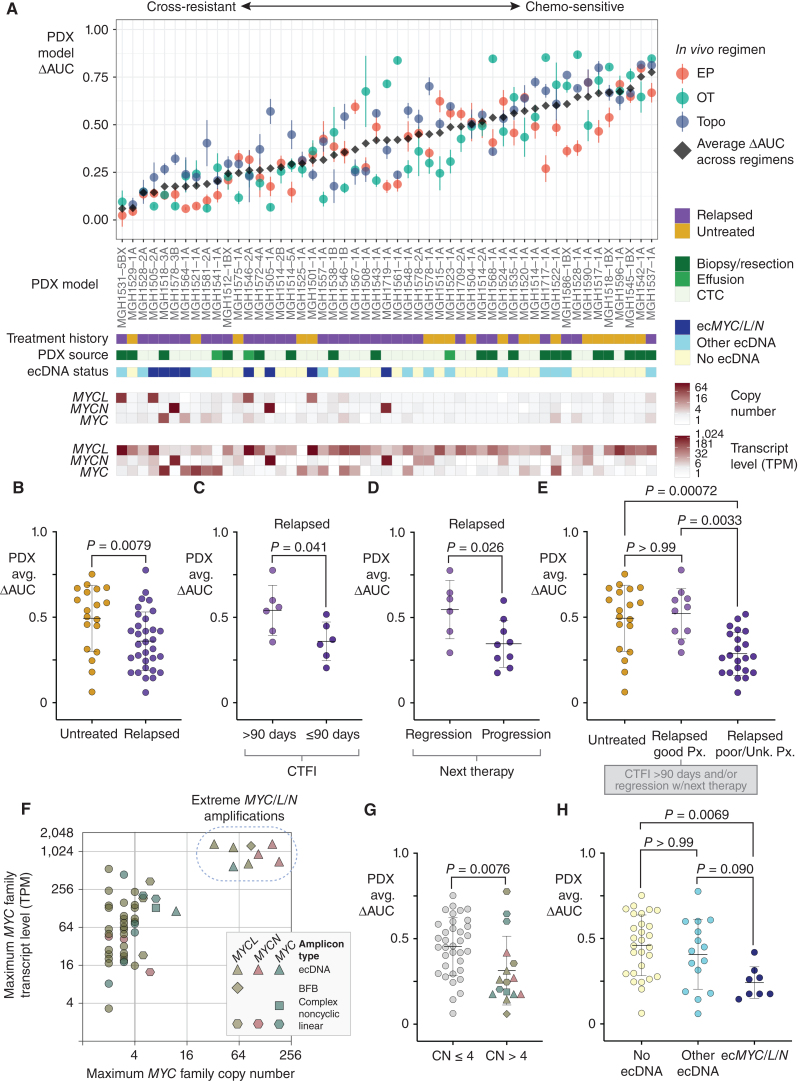
Figure 6.
ecDNA amplifications of MYC paralogs are recurrent in cross-resistant PDX models derived from patients with relapsed SCLC. A, Integrated clinical–functional–molecular landscape of MYC paralogs across the SCLC PDX panel. Top scatter plot: In vivo cross-resistance metrics. Models are arranged left-to-right by increasing the average of ΔAUC for EP, OT, and topotecan (ΔAUCavg). Error bars = SEM of ΔAUC for each regimen. Bars below (9 total): (1) Annotated clinical treatment history (chemo-naïve vs. post-relapse), (2) PDX source (CTC vs. biopsy vs. effusion), (3) ecDNA status (ecMYC/L/N vs. other ecDNA vs. no ecDNA), (4–6) MYC paralog copy numbers, and (7–9) MYC paralog transcript levels. B–E, Performance of PDX ΔAUCavg as a metric of clinical cross-resistance by comparison with patient clinical histories. B, Comparison of ΔAUCavg of PDX models derived from patients with untreated vs. relapsed SCLC. C, Comparison of ΔAUCavg of PDX models derived from patients after first-line therapy with CTFI > 90 days (platinum-sensitive) vs. ≤ 90 days (platinum-resistant). D, Comparison of ΔAUCavg of PDX models derived from patients with relapsed SCLC who responded to the next line of chemotherapy after model derivation vs. those who received chemotherapy but did not have significant tumor regression. E, Refinement of Fig. 6B to compare PDX ΔAUCavg between models derived from untreated patients, relapsed patients with positive prognostic features described in C and D (platinum sensitivity and response to next therapy), or relapsed patients with unknown or negative prognostic features. F, Comparison of transcript level vs. copy number for the MYC paralog with the highest expression level in each model. Amplicon type is annotated. Solid vertical line at CN = 4, beyond which we classify MYC paralogs as amplified. Models with > 30-copy amplifications form a clear separate cluster (“extreme amplifications,” dashed circle). G, Comparison of ΔAUCavg between models with or without MYC paralog amplifications (CN > 4), with annotation of gene and amplification structure. H, Comparison of ΔAUCavg between models without ecDNAs and models with ecDNAs with or without MYC paralogs. Statistical tests: Mann–Whitney test P values for B–D and G, and Kruskal–Wallis test P values for E and H. Created with BioRender.com.