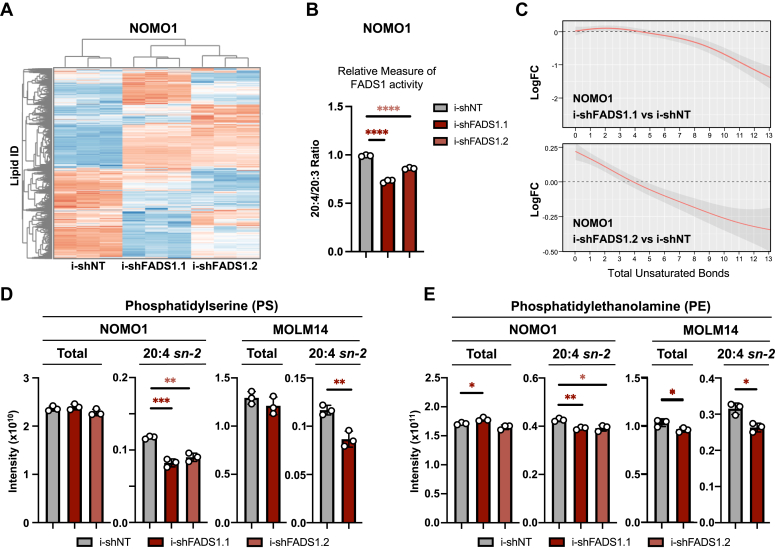
Figure 4.
FADS1 inhibition selectively depletes 20:4 fatty acid in phospholipids.A, hierarchical clustering heatmap of MS-based untargeted lipidomics analysis of NOMO1 cells expressing i-shNT, i-shFADS1.1, or i-shFADS1.2 at 40 h post doxycycline induction (n = 3 replicates per group). B, ratios of total signals from phospholipids containing 20:4 or 20:3 FA in the sn-2 position (∗∗∗∗p < 0.0001). C, analysis of fatty acid composition for i-shFADS1 versus control i-shNT cells based on the total MS signal (lipid abundance) with the specified total degree of unsaturation. Top panel, represents the comparison between i-shFADS1.1 versus control i-shNT and the bottom panel represents the comparison between i-shFADS1.2 versus control i-shNT. D and E, both total and 20:4, sn-2 levels of (D) phosphatidylserine (PS) (NOMO1 –i-shNT versus i-shFADS1.1: 20:4 sn-2 PS, ∗∗∗p = 0.0005; i-shNT versus i-shFADS1.2: 20:4 sn-2 PS, ∗∗p = 0.0015; and MOLM14 – i-shNT versus i-shFADS1.1: 20:4 sn-2 PS, ∗∗p = 0.0059) and (E) phosphatidylethanolamine (PE) were determined by MS-based untargeted NOMO1 cells expressing i-shNT, i-shFADS1.1, or i-shFADS1.2 and MOLM14 cells expressing i-shNT or i-shFADS1.1 at 40 h, post-DOX treatment (NOMO1 – i-shNT versus i-shFADS1.1: total PE, ∗p = 0.0284; and 20:4 sn-2 PE, ∗∗p = 0.0023; and i-shNT versus i-shFADS1.2: 20:4 sn-2 PE, ∗p = 0.015; MOLM14 – i-shNT versus i-shFADS1.1, 20:4 total PE, ∗p = 0.045; i-shNT versus i-shFADS1.1, 20:4 sn-2 PE, ∗p = 0.0107). Dots represent individual data points and error bars represent SD. FADS1, fatty acid desaturase 1; shFADS1, shRNAs that reduce Fads1 protein expression; shNT, nontargeting control shRNA.