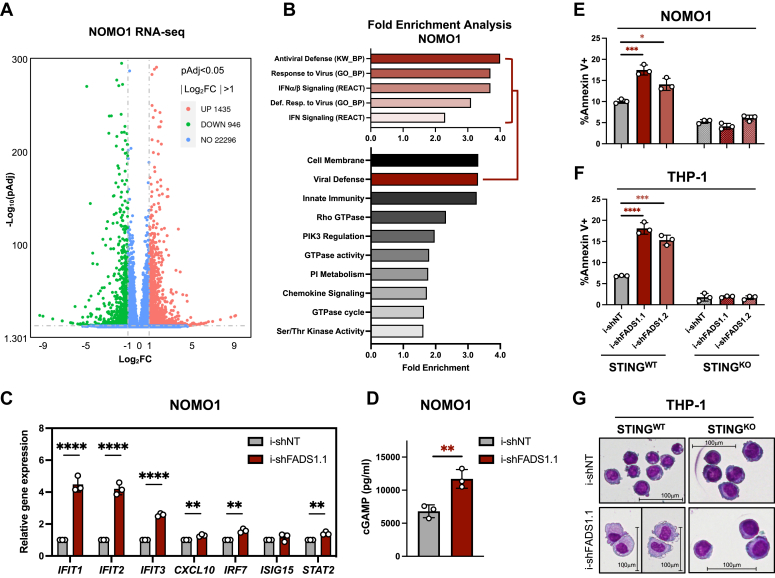
Figure 5.
FADS1 inhibition drives STING-mediated AML cell maturation and death.A, volcano plot of differentially expressed genes from a RNA-seq analysis of NOMO1 cells expressing i-shNT or i-shFADS1.1 and treated with DOX for 40 h. B, bottom panel, pathway enrichment analysis using the DAVID functional annotation tool and included GO_CC, GO_BP, GO_MF, UP_KW biological processes, KEGG, and reactome pathways. Top panel, pathway enrichment breakdown of “Viral Defense” cluster as depicted in the bottom panel. C, quantitative polymerase chain reaction analysis of the specified genes in i-shNT-, i-shFADS1.1-expressing NOMO1 cells at 40 h post doxycycline induction (i-shNT versus i-shFADS1.1: IFIT1, ∗∗∗p = 0.0002; IFIT2, ∗∗∗p = 0.0001; IFIT3, ∗∗∗∗p < 0.0001; CXCL10, ∗∗0.0024; IRF7, ∗∗p = 0.0012; STAT2, ∗∗p = 0.0025). D, cytoplasmic extracts from NOMO1-expressing i-shNT or i-shFADS1.1 and treated with DOX 40 h earlier were subjected to ELISA to detect cGAMP levels (p = 0.0078). E, NOMO1-STINGWT (i.e., WT). and NOMO1-STINGKO cells expressing i-shNT, i-shFADS1.1, or i-shFADS1.2 were analyzed for % annexin V+ (i-shNT versus i-shFADS1.1, ∗∗∗p = 0.006; i-shNT versus i-shFADS1.2, ∗p = 0.0104) using flow cytometry 4 days post-DOX induction. F, THP-1-STINGWT (i.e., WT). and THP-1-STINGKO cells expressing i-shNT, i-shFADS1.1, or i-shFADS1.2 were analyzed for % annexin V+ (i-shNT versus i-shFADS1.1, ∗∗∗p = 0.0001; i-shNT versus i-shFADS1.2, ∗∗∗p = 0.0002) using flow cytometry 5 days post-DOX induction. G, Wright-Giemsa staining of THP-1-STINGWT and THP-1-STINGKO cells expressing i-shNT or i-shFADS1.1, or i-shFADS1.2 shRNA at 5 days post doxycycline induction (20× magnification). Dots represent individual data points and error bars represent SD. AML, acute myeloid leukemia; BM, bone marrow; cGAMP, 2′3′-cGMP-AMP; FADS1, fatty acid desaturase 1; shNT, nontargeting control shRNA; STING, stimulator of interferon genes.