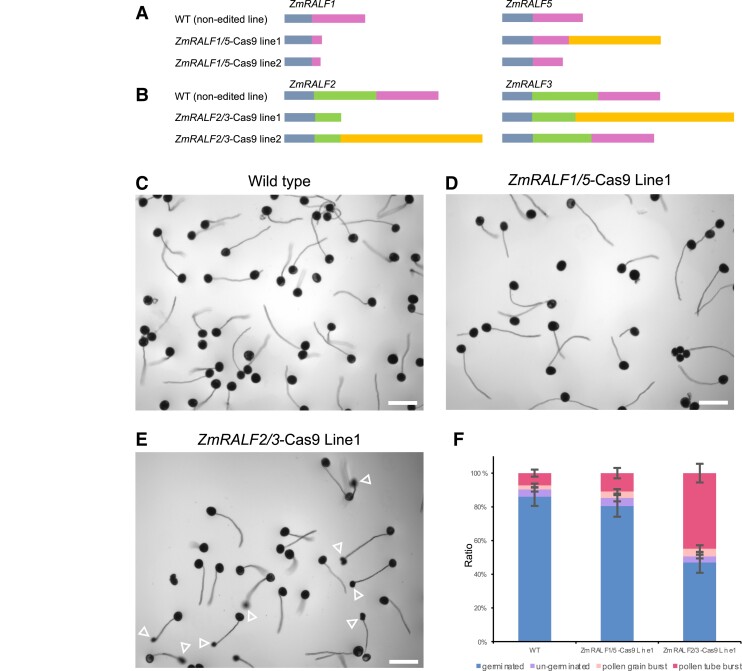
Figure 4.
Editing pattern and phenotypic analysis of ZmRALF1/5-Cas9 and ZmRALF2/3-Cas9 mutant lines. A) Genome editing pattern of ZmRALF1 and ZmRALF5 in mutant lines indicated. Blue-gray boxes show N-terminal signal peptides and pink boxes predicted mature peptides. Orange box indicates nonsense sequence. B) Genome editing pattern of ZmRALF2 and ZmRALF3 in mutant lines indicated. Color code of boxes as in A). Green box additionally shows the cleaved pro-peptide sequence (see also Fig. 1B; Supplementary Fig. S2A). C to E) In vitro pollen germination of WT (non-edited transgenic line) in C), ZmRALF1/5-Cas9 Line1 D), and ZmRALF2/3-Cas9 Line1 E), respectively. Pollen grains were germinated on PGM for 60 min. Arrowheads in E) point toward burst pollen tubes. Scale bars are 200 μm. F) Quantification of pollen burst ratios in WT (non-edited transgenic line), ZmRALF1/5-Cas9 Line1 and ZmRALF2/3-Cas9 Line1, respectively. n > 200 in every plant from each line. The data are presented as the mean value ± Sds from 3 independent pollen germination experiments.