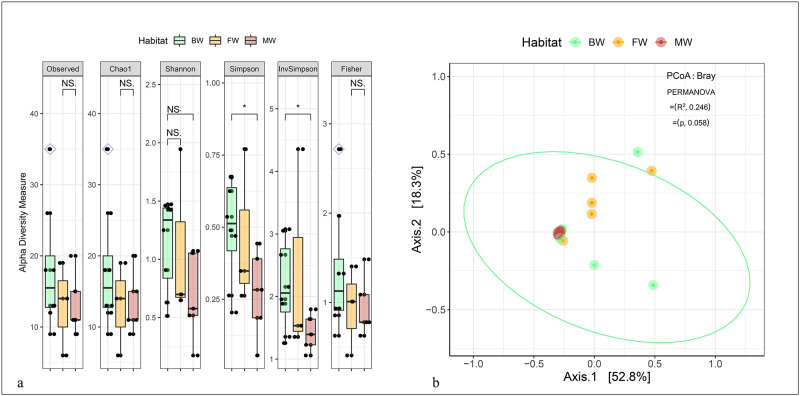
Fig 1. Bacteriome diversity in hilsa fish.
(a) Within subject (Alpha) diversity measure. Observed, Chao1, Shannon, Simpson, InvSimpson and Fisher indices estimated alpha diversity in hilsa fish samples according to host habitat i.e., freshwater (FW), brackish water (BW) and marine water (MW). The within sample diversity are plotted on boxplots and comparisons are made with pairwise Wilcoxon rank sum tests. Significance level (p-value) 0.01 and 0.05 are represented by the symbols "**", and "*", respectively. (b) Between subject (Beta) diversity measure according to host habitat i.e., FW, BW and MW. Bacterial beta diversity was calculated using Bray-Curtis dissimilarity distance method, and visualized on principal coordinate analysis (PCoA) plots. The samples are coloured according to host habitat (e.g., FW: cheese orange, BW: dragon green and MW: cherry red) and joined with the respective ellipses. Pairwise comparisons on a distance matrix using PERMANOVA test under reduced model shows significant bacterial community differences across the habitats (p = 0.05, R2 = 0.246). NS refers to non-significant.