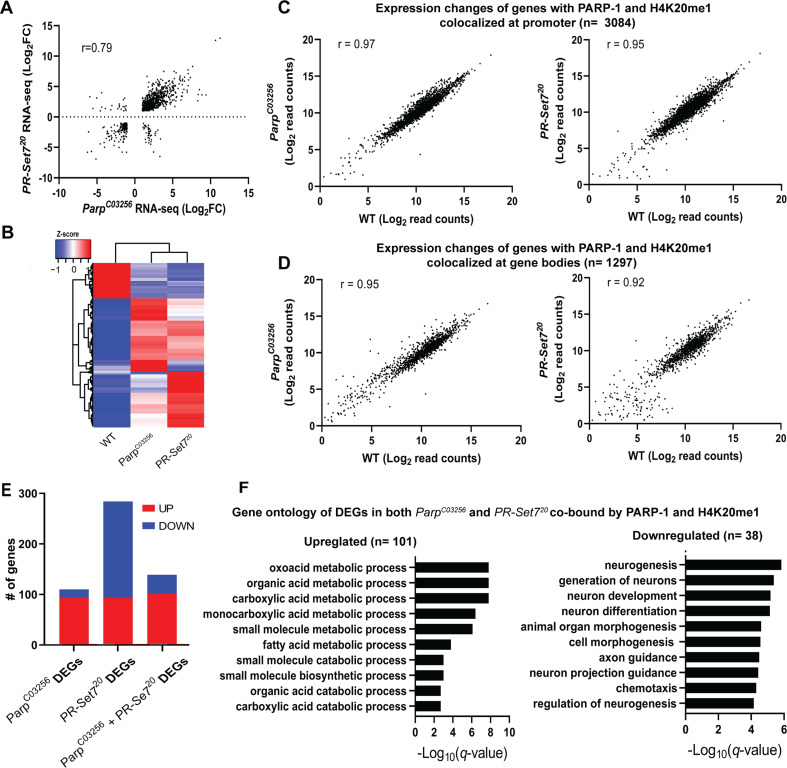
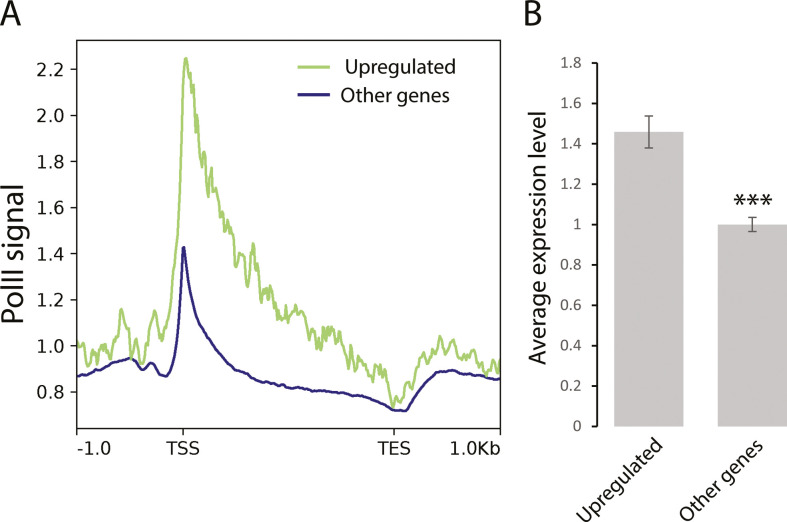
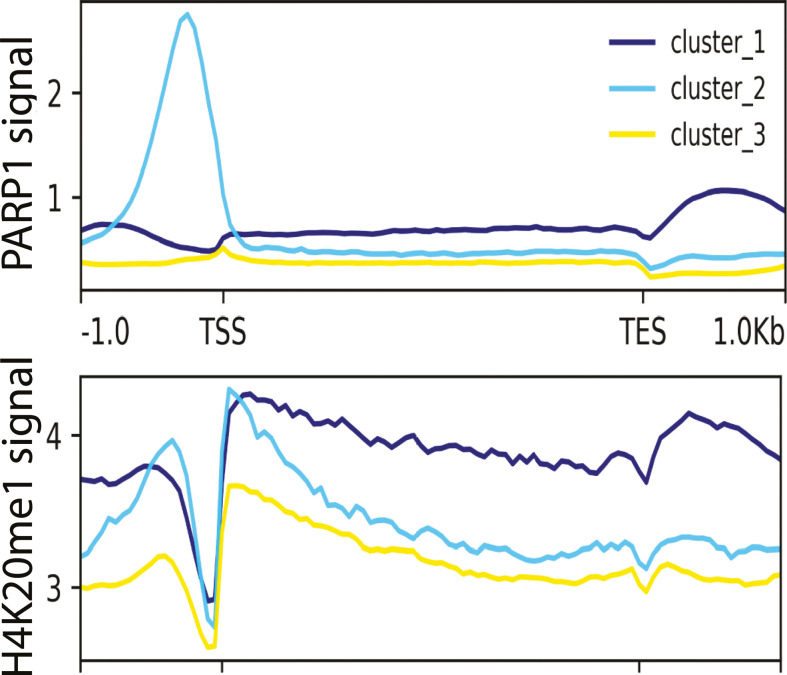
Figure 3. PARP-1 and H4K20me1 are required for the repression of metabolic genes and activation of developmental genes at co-enriched genes.
(A) Scatterplot plot showing correlation of differentially expressed genes (DEGs) in parp-1C03256 and pr-set720 (Pearson’s r=0.79). (B) Heatmap showing the normalized read counts of DEGs in both parp-1C03256 and pr-set720. Normalized read counts are shown as row z-scores. (C–D) Dot plots showing transcriptional changes of genes co-enriched with PARP-1 and H4K20me1 in parp-1C03256 and pr-set720 compared to WT at promoters (C) and gene bodies (D). (E) Summary of DEGs in parp-1C03256 and pr-set720 and both mutants that were co-enriched with PARP-1 and H4K20me1. (F) Gene ontology of upregulated (left) and downregulated (right) DEGs in both parp-1C03256 and pr-set720 mutants that were co-enriched with PARP-1 and H4K20me1.
Figure 3—figure supplement 1. Validation of pr-set720 mutant.
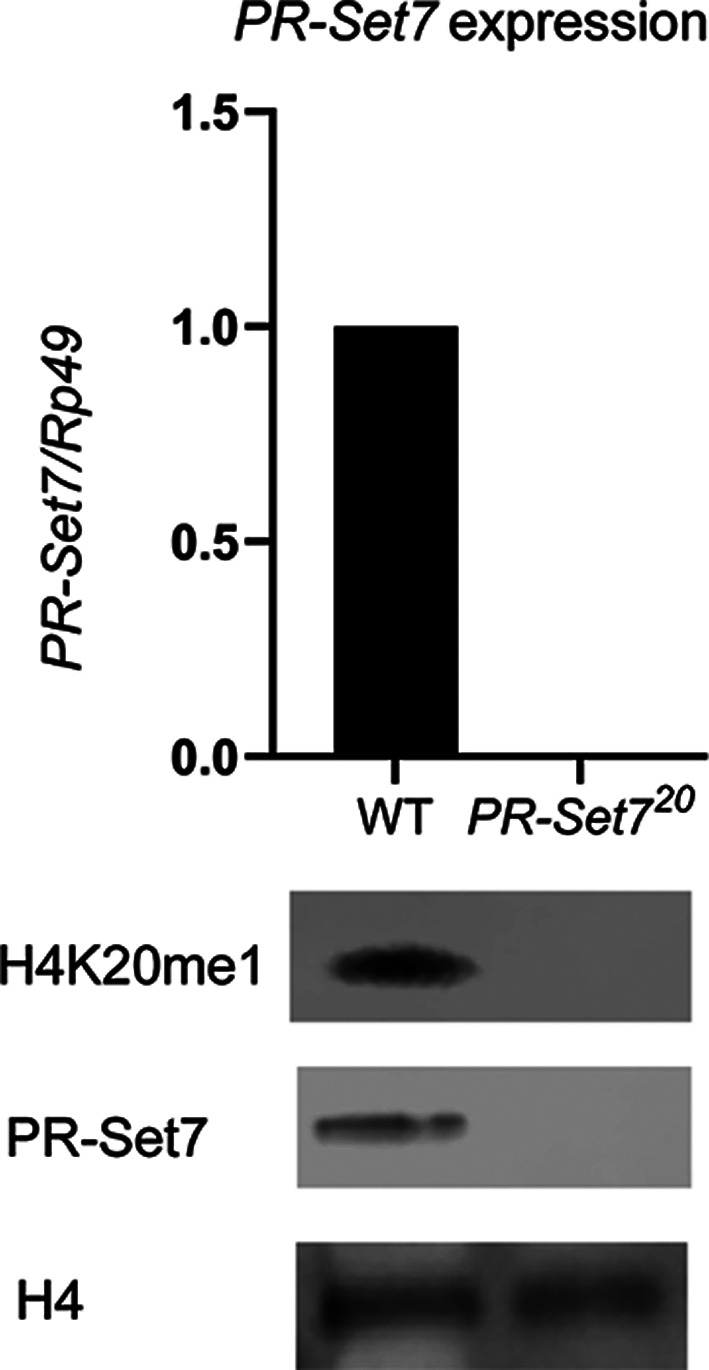
Figure 3—figure supplement 2. PR-SET7 expression level is not affected in parp-1C03256 mutant.
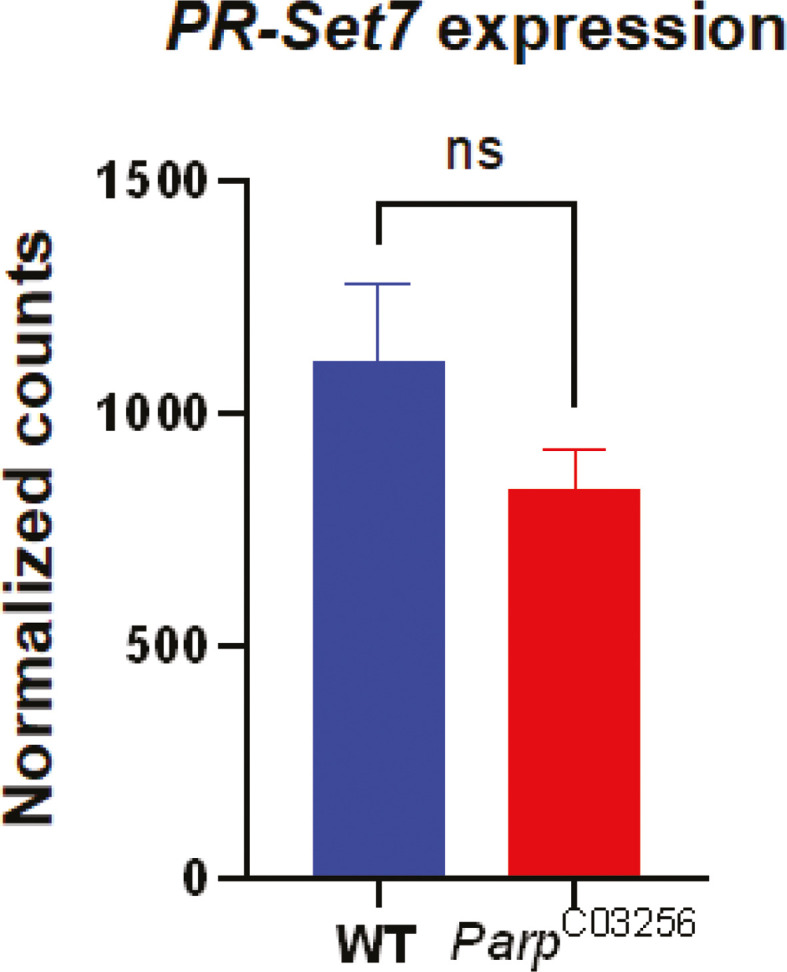
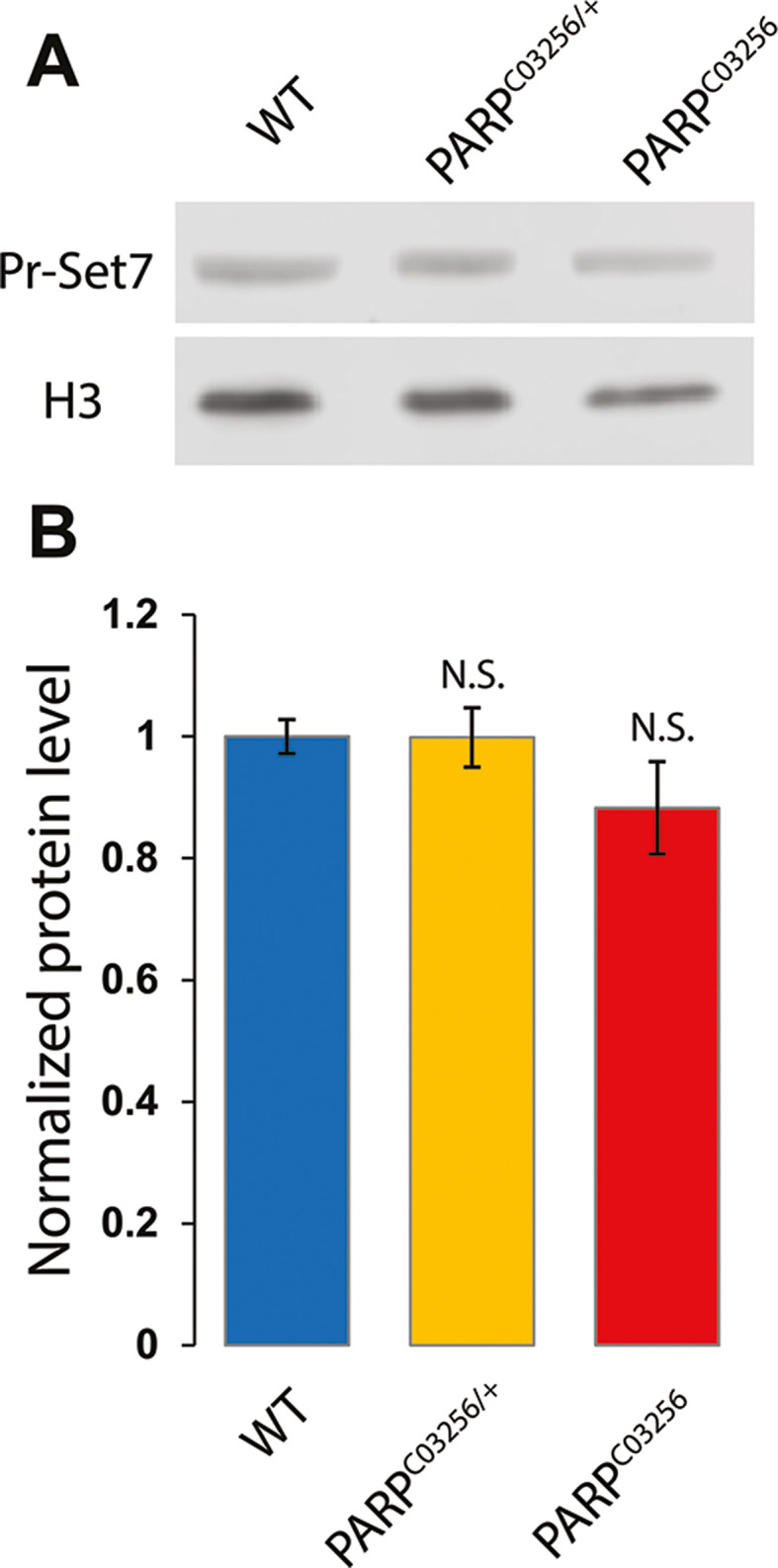
Figure 3—figure supplement 3. PR-SET7 protein level is not affected in parp-1C03256 mutant.