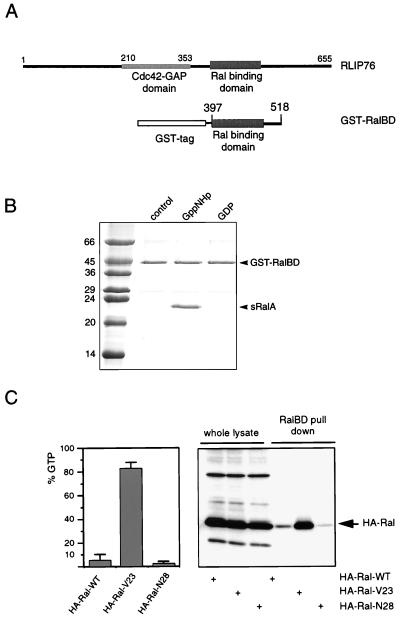
FIG. 1.
A GST fusion protein of the RalBD of RLIP76 binds specifically to the activated form of Ral. (A) Schematic representation of RLIP76, indicating the position of the RalBD of RLIP76 that was used to generate the GST-RalBD. (B) In vitro analysis of the interaction between GST-RalBD and sRalA. A 3μM concentration of GST-RalBD was precoupled to glutathione-Sepharose beads and washed. A 4 μM concentration of a C-terminally truncated sRalA loaded with GDP or Gpp(NH)p (a nonhydrolyzable GTP analog) was incubated with the precoupled beads. After 1 h of incubation at 4°C, beads were washed three times and resuspended in 80 μl of sample buffer, of which 10 μl was loaded onto an SDS-PAGE gel. After separation, the proteins were stained by Coomassie brilliant blue. The numbers are molecular masses of the marker proteins, in kilodaltons. (C) Analysis of GTP levels of HA-Ral mutants by different methods. The left panel shows the percentage of GTP bound to ectopically expressed wild-type HA-Ral (HA-Ral-WT), activated HA-Ral (HA-RalV23), and inactive HA-Ral (HA-RalN28) in serum-starved Cos7 cells. Error bars indicate the variations between two independent experiments. The right panel shows the detection of HA-RalGTP in a GST-RalBD pull-down experiment. Therefore, the transfected Cos7 cells were serum starved, lysed, and incubated with 15 μg of GST-RalBD precoupled to glutathione beads to recover GTP-bound Ral. Beads were washed four times, and collected Ral was identified by Western analysis with an anti-HA (12CA5) monoclonal antibody (lanes 4 to 6). The first three lanes show the levels of the transfected HA-Ral mutants in the whole lysates. In these lanes, 5% of the lysate used for the pull-down experiment was loaded onto the gel.