Figure 2.
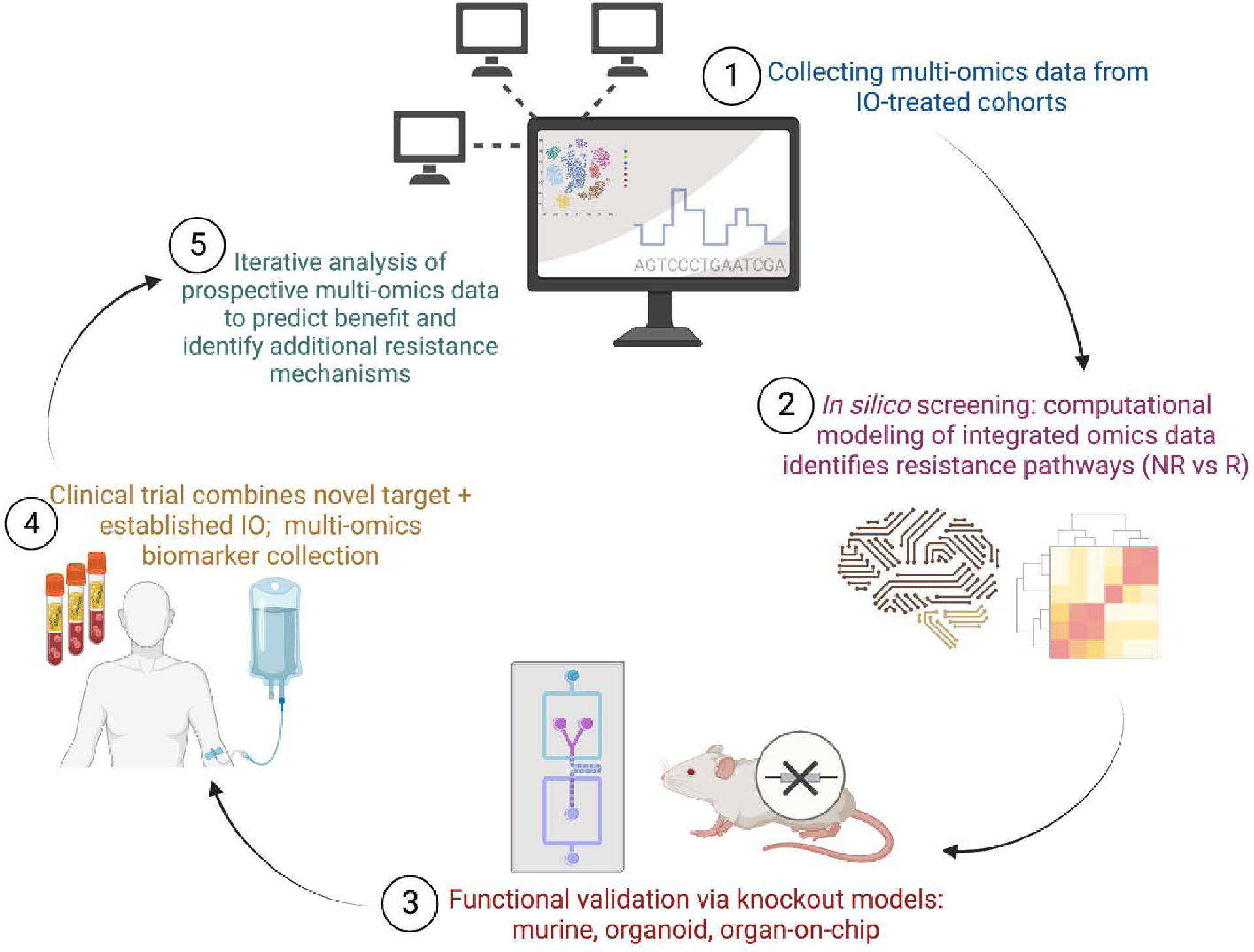
Framework for utilizing multi-omics data to advance immunotherapy combination strategies. An iterative cycle of in silico screening, functional validation, and clinical trial biomarker collection can be used to identify novel resistance pathways, develop predictive models of benefit, and highlight additional resistance mechanisms.
In step 1, multi-omics and/or image data is collected from clinically annotated or biologically distinct patient cohorts, using bulk RNAseq, single-cell RNAseq, multispectral imaging, spatial profiling, among others. These data are computationally integrated and compared between IO-non-responders and IO-responders along with non-T cell-inflamed versus T cell-inflamed tumors samples. This in silico screening identifies candidate genes and molecular pathways that may be associated with IO-resistance and/or immune exclusion for therapeutic target nomination (step 2). In step 3, functional assays are used to experimentally validate the immune-exclusive role of these candidate genes/pathways and assess novel inhibitors and other agents. Based on these preclinical data, early phase clinical trials combine novel pathway agents with established IO therapies; clinical outcome along with translational biomarker data is collected (step 4). In returning to the start of the cycle (step 5), multi-omics biomarker data from these novel clinical trials are used to identify predictors of response as well as additional genes/pathways tied to therapy resistance.
IO, immuno-oncology; NR, non-responder; R, responder
Figure made with BioRender
