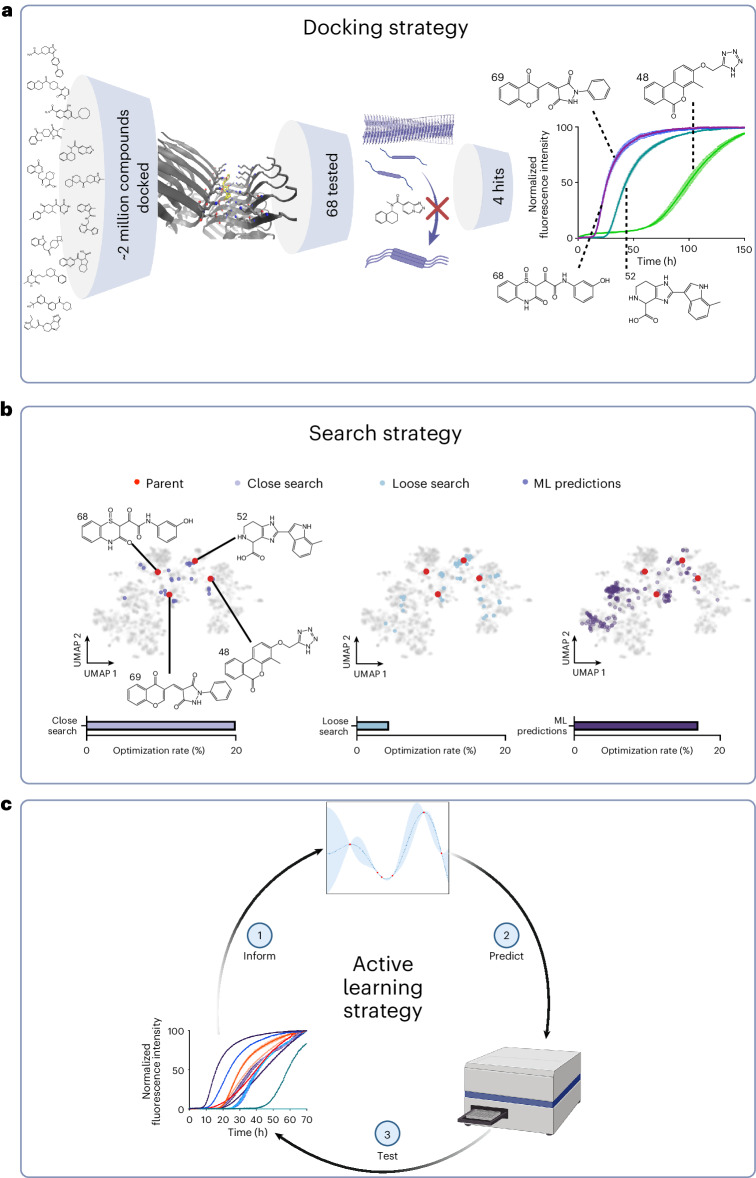
Fig. 1. Illustration of the three stages of exploration of the chemical space described in this work.
a, From 68 molecules predicted to have good binding via docking simulations, we initially identified 4 active molecules (the ‘docking set’) by experimental testing23. These four molecules increase the t1/2 of αS aggregation. b, We then performed a close Tanimoto similarity search around the four parent compounds in chemical space. We selected molecules with Tanimoto similarity cutoff >0.5 (the ‘close similarity docking set’) followed by a loose similarity search with Tanimoto similarity cutoff >0.4 (the ‘loose similarity docking set’). A machine learning method was then applied using the observed data to predict potent molecules from a compound library derived from the ZINC database with Tanimoto similarity >0.3 to the parent structures (the ‘evaluation set’). c, Successive iterations of prediction and experimental testing yielded higher optimization rates (defined as the percentage of molecules increasing the normalized half time of aggregation above 2), and molecules with higher potency on average than those identified in the previous similarity searches. Validation experiments were also carried out on the potent molecules identified.