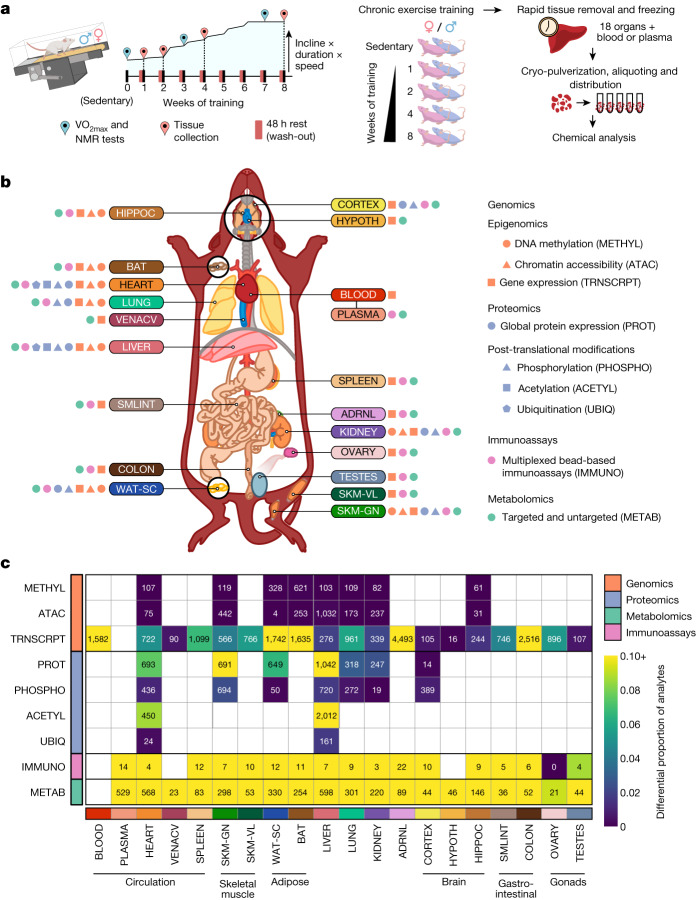
Fig. 1. Summary of the study design and multi-omics dataset.
a, Experimental design and tissue sample processing. Inbred Fischer 344 rats were subjected to a progressive treadmill training protocol. Tissues were collected from male and female animals that remained sedentary or completed 1, 2, 4 or 8 weeks of endurance exercise training. For trained animals, samples were collected 48 h after their last exercise bout (red pins). b, Summary of molecular datasets included in this study. Up to nine data types (omes) were generated for blood, plasma, and 18 solid tissues, per animal: ACETYL: acetylproteomics; protein site acetylation; ATAC, chromatin accessibility, ATAC-seq data; IMMUNO, multiplexed immunoassays; METAB, metabolomics and lipidomics; METHYL, DNA methylation, RRBS data; PHOSPHO, phosphoproteomics; protein site phosphorylation; PROT, global proteomics; protein abundance; TRNSCRPT, transcriptomics, RNA-seq data; UBIQ, ubiquitylome, protein site ubiquitination. Tissue labels indicate the location, colour code, and abbreviation for each tissue used throughout this study: ADRNL, adrenal gland; BAT, brown adipose tissue; BLOOD, whole blood, blood RNA; COLON, colon; CORTEX, cerebral cortex; HEART, heart; HIPPOC, hippocampus; HYPOTH, hypothalamus; KIDNEY, kidney; LIVER, liver; LUNG, lung; OVARY, ovaries; PLASMA, plasma; SKM-GN, gastrocnemius (skeletal muscle); SKM-VL, vastus lateralis (skeletal muscle); SMLINT, small intestine; SPLEEN, spleen; TESTES, testes; VENACV, vena cava; WAT-SC, subcutaneous white adipose tissue. Icons next to each tissue label indicate the data types generated for that tissue. c, Number of training-regulated features at 5% FDR. Each cell represents results for a single tissue and data type. Colours indicate the proportion of measured features that are differential.