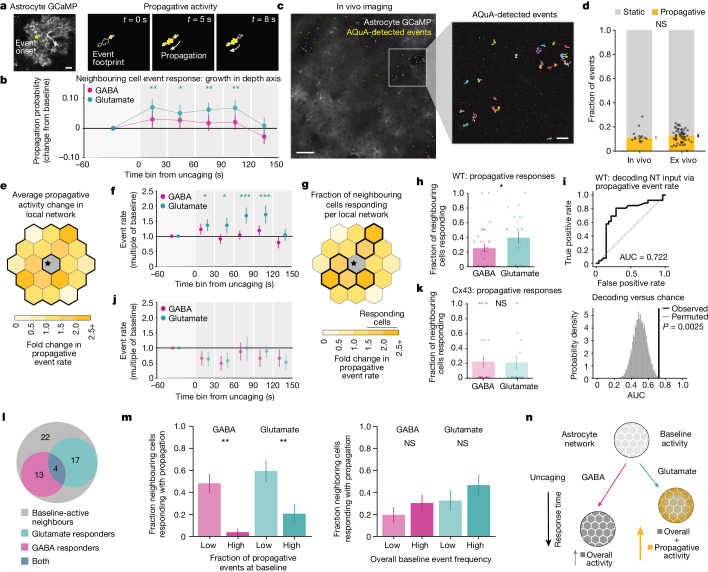
Fig. 4. Propagative activity distinguishes astrocyte network responses to GABA and glutamate.
a, Astrocytic GCaMP6f fluorescence with initial territory (left) and subsequent trajectory (right) of a propagating event in yellow. Outline: total event territory. b, Probability change of Ca2+ event growing in the depth axis (relative to pia) among all events from neighbouring cells after NT uncaging. Data shown as overall probability ± standard error (n = 142 cells, 28 FOV (GABA), 120 cells, 27 FOV (glutamate)). Two-sided P and q values by permutation testing (Supplementary Table 8). c, Two-photon image of in vivo astrocyte GCaMP6f in V1. Overlay: Ca2+ events from 90-s stationary period. d, Propagative event fraction in V1 during stationary wakefulness in vivo and baseline in acute V1 slices. Data shown by recording; median ± standard error by bootstrapping (n = 15 recordings, 5 mice (in vivo), 55 recordings, 4 mice (ex vivo)). Two-sided rank-sum test (P = 0.57). e,f,j, Schematic (e) and quantification of fold change in propagative event rate across neighbouring cells per FOV after NT uncaging in WT (f) or Cx43-floxed (j) slices. Data shown as median across FOVs ± standard error. One-sided P and q values by permutation testing (see Supplementary Tables 9 and 10). As in Fig. 3, directly stimulated astrocyte excluded from all figure analyses. g,h,k, Schematic (g) and quantification of fraction of neighboring cells per FOV with ≥50% propagative event rate increase (‘responding’) after NT uncaging in WT (h) or Cx43-floxed (k) slices. Data shown by FOV; mean ± sem (see Supplementary Table 9). Two-sided P values by permutation testing, P = 0.046 (WT), 1.0 (Cx43-floxed). i, Top: receiver operating characteristic curve decoding NT identity by thresholding relative propagative event rate change across all neighbouring cells per FOV. Bottom: observed area under the receiver operating characteristic curve (AUC) = 0.72 ± 0.077 (value ± bootstrapped standard error), compared to permuted distribution through permuting NT labels (P = 0.0025, n = 55 FOVs, one-sided). l, Neighbouring cell numbers responding to one or both NTs with propagative activity increases, among cells with baseline propagative activity (n = 56 cells, 24 paired recordings, 7 slices, 4 mice). Permutation testing measures of correlation (two-sided Spearman ρ, P = 0.24) or overlap (one-sided Jaccard index, P = 0.96) between GABA and glutamate responses. m, Fraction of neighbouring cells responding with propagative increases after NT uncaging, cells equally divided by low and high baseline activity features (split at 50th percentile). Baseline activity features: fraction of propagative events (left), overall event rate (right, see Extended Data Fig. 5e). Data shown as mean ± s.e.m. (see Supplementary Table 11). Response fractions for cells with ‘low’ and ‘high’ baseline fractions were compared by permuting cells’ baseline propagation fractions for GABA (P = 1.0 × 10–4) and glutamate (P = 0.0012); responses for cells with ‘low’ and ‘high’ overall baseline event rates were compared similarly (GABA: P = 0.25; glutamate: P = 0.25). n, Integrated model of astrocyte network responses. Astrocyte networks increase general Ca2+ with both NTs, and propagative activity specifically with glutamate. Network responses to glutamate are faster than those to GABA. b,f,h,j,k,m, error bars by hierarchical bootstrapping. b,f, *q < 0.05, **q < 0.01, ***q < 0.001, h,m, *P < 0.05, **P < 0.01. Scale bars, 10 μm (a,c (right)) and 50 μm (c (left)).