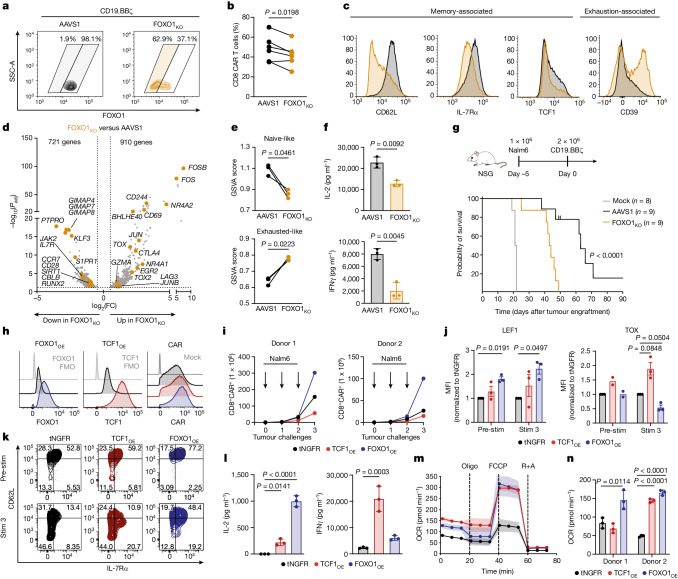
Fig. 1. FOXO1 is necessary and sufficient for memory and antitumour function in human CAR T cells.
a–g, CRISPR–Cas9 gene editing of AAVS1 (AAVS1) or FOXO1 (FOXO1KO) in CD19.BBζ CAR T cells. a–c, Flow cytometric analysis of FOXO1 knockout efficiency (a), percentage of CAR+ CD8+ cells at day 14 (b) and memory- and exhaustion-associated markers in CAR+CD8+ cells (c). Shaded areas in a represent gates used in phenotypic analyses. One representative donor is shown in a and c (n = 6 donors). d, Volcano plot of DEGs in CD62Llo FOXO1KO versus AAVS1 (Bonferroni-adjusted P < 0.05 with absolute log2-transformed fold change (abs(log2(FC)) > 0.5). e, GSVA using T cell gene signatures55. f, Cytokine secretion in response to Nalm6 leukaemia cells from one representative donor (n = 4 donors). g, Stress test Nalm6 xenograft model. Top, schematic. Bottom, survival curves of Nalm6-engrafted mice treated with mock T cells or gene-edited CD19.BBζ cells. Data show two donors tested in two independent experiments (n = 8 or 9 mice per group). Data in d and e include n = 3 donors. h–n, CAR T cells overexpressing truncated NGFR (tNGFR), TCF1-P2A-tNGFR (TCF1OE) or FOXO1-P2A-tNGFR (FOXO1OE). h, Flow cytometric analysis of FOXO1, TCF1 and CD19.28ζ expression from one representative donor (n = 8 donors). FMO, fluorescence minus one. i–k, Serial restimulation of CD19.BBζ cells with Nalm6. CD8+ CAR T cell expansion (i) and flow cytometric analysis of memory- and exhaustion-associated markers (j,k). j, Mean ± s.e.m. of normalized mean fluorescence intensity (MFI) (n = 2 or 3 donors). k, One representative donor (n = 4 donors). l, HA.28ζ cytokine secretion (day 13) in response to 143B osteosarcoma cells from one representative donor (n = 4 donors). m,n, HA.28ζ seahorse analysis (day 13) (n = 2 donors). m, Oxygen consumption rate (OCR) (mean ± s.d. of 11 technical replicates from one representative donor). Oligo, oligomycin; R+A, rotenone and antimycin. n, Spare respiratory capacity. Data in f,l,n are mean ± s.d. of three technical replicates. Statistical comparisons were performed using paired two-tailed Student’s t-test (b,e), two-sided Welch’s t-test (f), log-rank Mantel–Cox test (g) and repeated-measures one-way ANOVA with Geisser–Greenhouse correction (j) or one-way ANOVA with Dunnett’s test (l,n) .