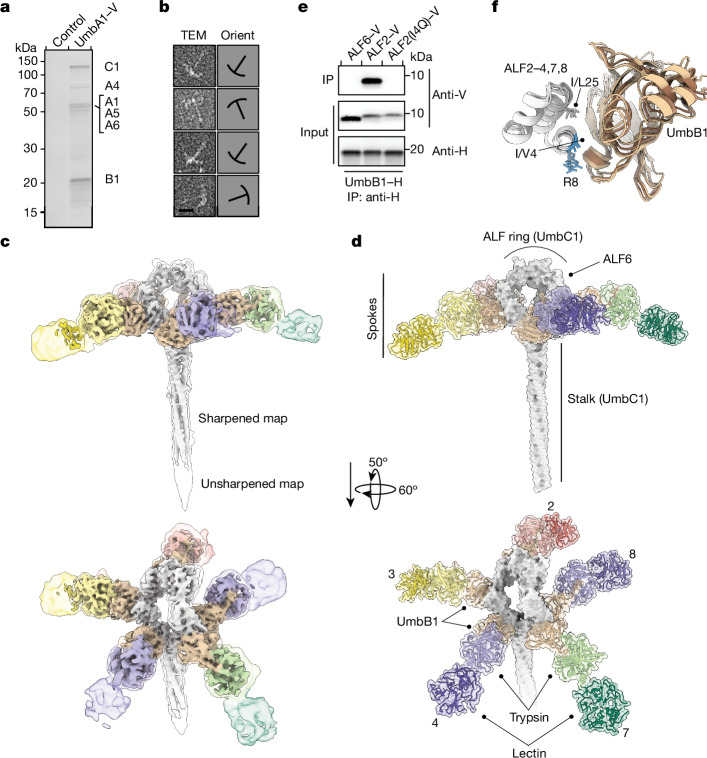
Fig. 3. Structure of the Umb1 particle.
a, Silver-stained SDS–PAGE analysis of the Umb1 protein complex precipitated from cultures of S. coelicolor with chromosomally encoded UmbA1–V. Control represents the equivalent precipitation from wild-type S. coelicolor. Experiment shown is representative of three independent replicates. b, Transmission EM analyses of negative-stained, purified Umb1 particles. Outlines indicating particle orientation (Orient) are shown on the right. Complete micrographs are provided in Supplementary Fig. 2a. c, Cryo-EM maps of the Umb1 particle at 4.3 Å unsharpened (transparent, 0.43 binarization threshold) and sharpened (opaque, 1.16). Maps are coloured based on model subunit proximity. Regions encompassing UmbB1 and UmbC1 are coloured as in Fig. 2, and regions encompassing UmbA1 are coloured differently in each spoke to highlight the complement of UmbA proteins that associate with the Umb1 particle. d, Depiction of the Umb1 particle. The depiction represents the model derived from our cryo-EM data, with the exception that the lectin domains present in spokes 2, 4 and 7 are shown but were not included in the deposited model owing to their weak density. The C-terminal domains of UmbC1 were not resolved in our structure and are not included in the depiction. Spoke numbers correspond to the interacting ALF repeat of UmbC1. ALF1, ALF5 and ALF6 (indicated) do not interact with UmbB1. e, WB analysis of IP experiments between UmbB1 and the indicated native and variant ALF repeats. Protein tags are indicated. f, Superimposition of UmbB1-binding ALF repeats (white) in complex with their corresponding UmbB1 protomer (brown shades) extracted from the Umb1 particle model. Conserved residues at the ALF–UmbB1 interface are shown, with several side chains truncated at Cβ as in the model. Those not conserved in ALF6 are coloured blue.