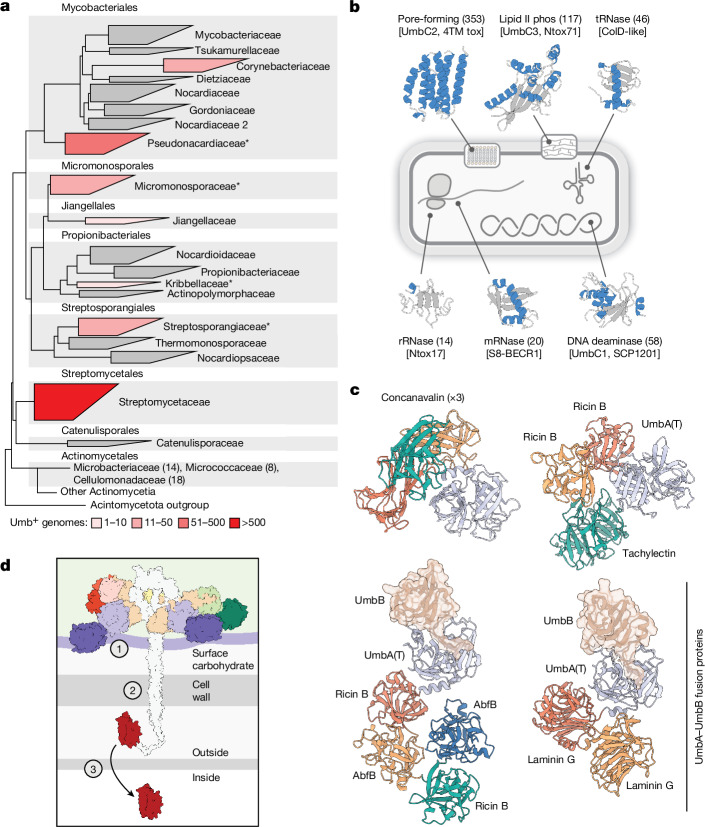
Fig. 5. Phylogenetic distribution and functional diversity of Umb proteins.
a, Phylogenetic tree of orders and families within Actinomycetia, coloured to indicate the number of genomes positive for Umb toxin particle loci. Within Actinomycetales, only those families containing umb loci are listed, with the number of umb-containing genomes in parentheses. Asterisks indicate families for which representative umb loci are shown in Supplementary Fig. 5. The width of boxes for each family is proportional to the number of species it contains. b, Schematic indicating the predicted molecular targets of select toxin domains commonly found in UmbC proteins and representative models for the domains generated using AlphaFold. Models coloured by secondary structure (blue, α-helices; grey, loops and β-strands). Numbers in parentheses indicate the number of UmbC proteins we detected carrying the indicated toxin domain. Toxin family names are provided in brackets and in Supplementary Table 2. c, Predicted structural models of example UmbA proteins selected by virtue of containing multiple distinct or repeated lectin domains (top) or fusions between UmbA and UmbB proteins (bottom). The UmbB domains of bifunctional UmbAB proteins are shown in transparent surface representation and in the same orientation to highlight their conserved interaction with the major cleft of the trypsin-like domain. Concanavalin, UAL-Con-1; Ricin B, Ricin_B_Lectin (Supplementary Table 4). d, Model for the intoxication of target cells by Umb toxins, with outstanding questions highlighted. These include the identity of receptor (or receptors) on target cells and the involvement of the lectin domains in mediating binding (1), the role of the stalk in toxin delivery (2) and the mechanism of toxin translocation into target cells (3).