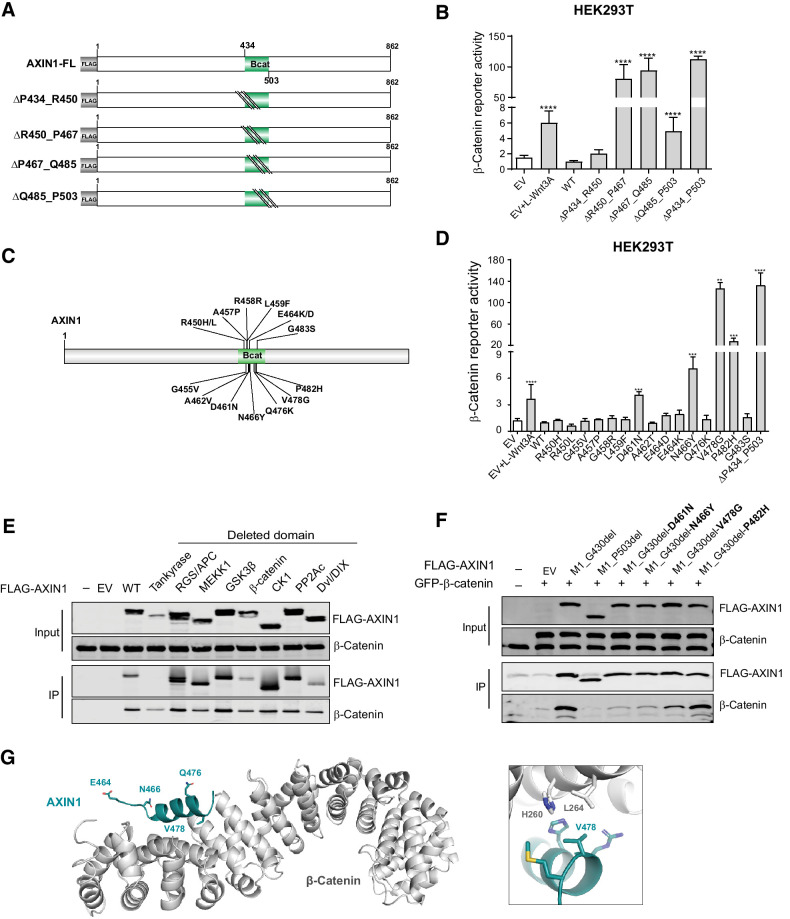
Figure 4.
Missense variants in the β-catenin–binding domain mostly retain functionality. Tumor-associated missense variants within the β-catenin–binding domain were tested for their effect on β-catenin signaling regulation. A, Schematic diagram of the indicated sequential amino acid deletions that were generated in the AXIN1 expression plasmid. B, A β-catenin reporter assay was performed to show the defect in β-catenin regulation associated with the variants shown in A. C, Schematic diagram depicting the 15 selected tumor-associated missense variants within the R450_P503 domain. D, A β-catenin reporter assay to determine the defect in β-catenin regulation for the missense variants shown in C. Reporter activities are depicted as WRE/CMV-Renilla ratios (mean ± SD; three independent experiments), in which the value obtained for the empty vector (EV) was arbitrarily set to 1. E, Immunoprecipitation experiment with FLAG-tagged AXIN1 variants to identify which domain deletions affect endogenous β-catenin binding. F, Immunoprecipitation experiment with depicted FLAG-tagged AXIN1 missense variants to determine β-catenin–binding capacity. Deletion of the N-terminal half of AXIN1 up to the β-catenin domain (M1_G430del) was used to allow a better evaluation. A larger deletion (M1_P503del) also removing the β-catenin domain was used as negative control. Cotransfected GFP–β-catenin and endogenous β-catenin were detected using a β-catenin antibody. In all IP experiments, transfection with EV and nontransfected cells was used as negative controls. Data are shown as mean ± SD. G, Cartoon representation of β-catenin (gray) with bound AXIN1 α-helix (deep teal). Side chains of amino acid residues that are mutated in this study are indicated in stick representation, with nitrogen atoms (blue) and oxygen (red). Right, close-up of the region around AXIN1 residue Valine 478. Statistical significance for all experiments was analyzed using a Mann–Whitney test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.