Figure 1.
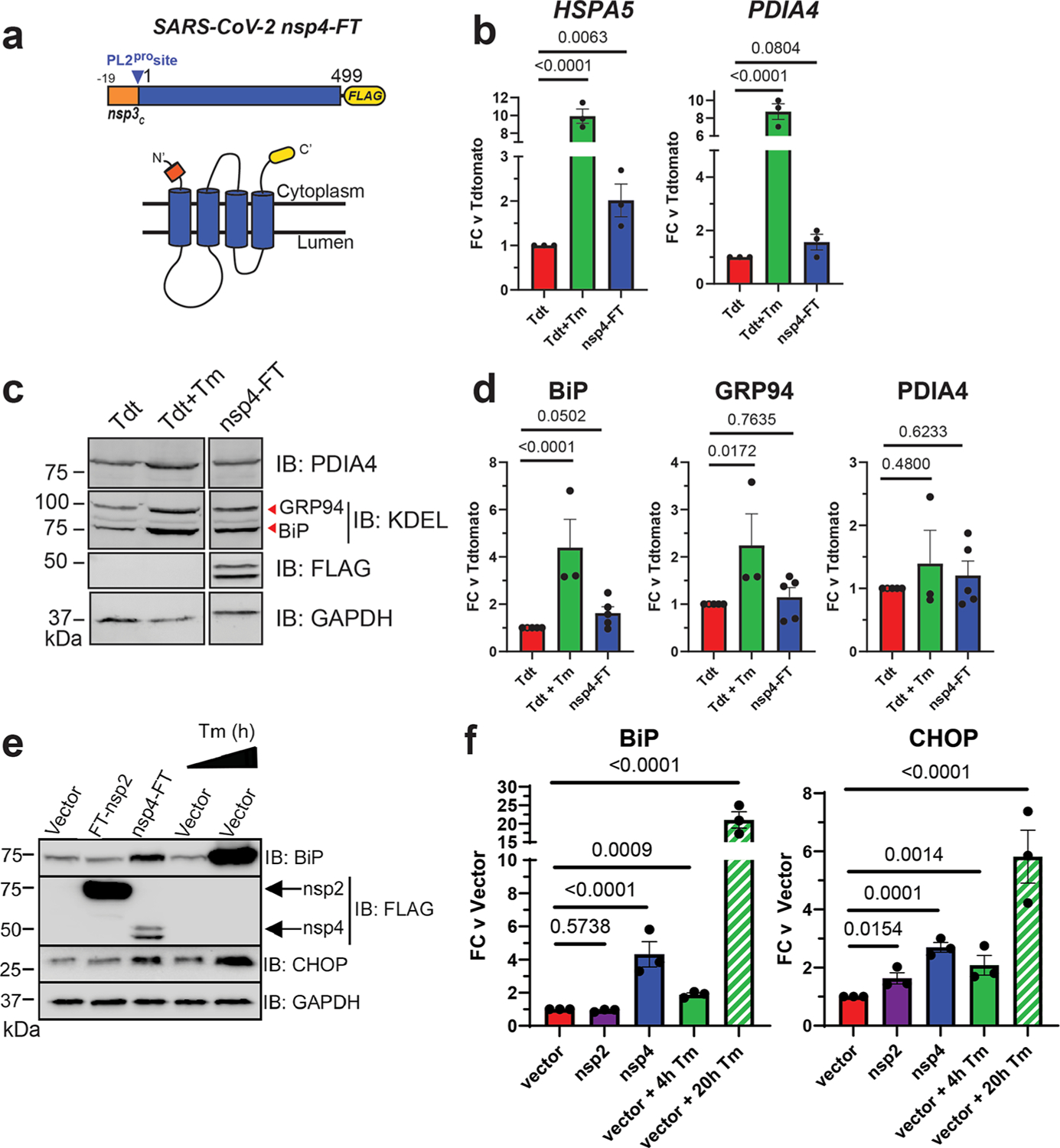
SARS-CoV-2 nsp4 upregulates the expression of UPR reporter proteins. (a) SARS-CoV-2 nsp4 expression construct and membrane topology, containing a C-terminal FLAG-tag (nsp4-FT) and an N-terminal segment of the last 19 C-terminal amino acids in nsp3 for optimal membrane insertion.21 Construct contains the native PL2pro protease cleavage site. (b) RT-qPCR of ATF6 pathway activation reporters HSPA5 and PDIA4 in the presence of SARS-CoV-2 nsp4-FT, normalized to GAPDH transcripts and compared to basal levels (GFP). Treatment with 1 μg/mL Tunacimycin (Tm) for 6 h was used as a positive control. n = 3, mean ± SEM, one-way ANOVA with Benjamini, Krieger, and Yekutieli multiple testing correction, p < 0.05 considered statistically significant. (c) Representative western blot of ATF6 protein markers (PDIA4, GRP94, BiP) in the presence of Tdtomato (Tdt), SARS-CoV-2 nsp4-FT, or treatment with 1 μg/mL Tm (16 h), with GAPDH as a housekeeping gene for loading control. Displayed blot sections are from the same blot image and exposure settings (see Supplemental Figure S1 for all quantified, original blots). (d) Quantification of Tdtomato (Tdt), and nsp4-FT in western blots in panel (c), normalized to GAPDH band intensities, and compared to basal levels (Tdt). n = 3–5, mean ± SEM, one-way ANOVA with Benjamini, Krieger, and Yekutieli multiple testing correction, p < 0.05 considered statistically significant. (e) Representative western blot of CHOP and BiP in the presence of SARS-CoV-2 FT-nsp2, nsp4-FT, or treatment with 5 μg/mL Tm for short (4h) or long (20h) time points. See Supplemental Figure S2 for all quantified, original blots. (f) Quantification of western blots in panel (e), normalized to GAPDH band intensities and compared to basal control (vector). n = 3, mean ± SEM. One-way ANOVA with Benjamini, Krieger, and Yekutieli multiple testing correction, p < 0.05 considered statistically significant.
