Figure 3.
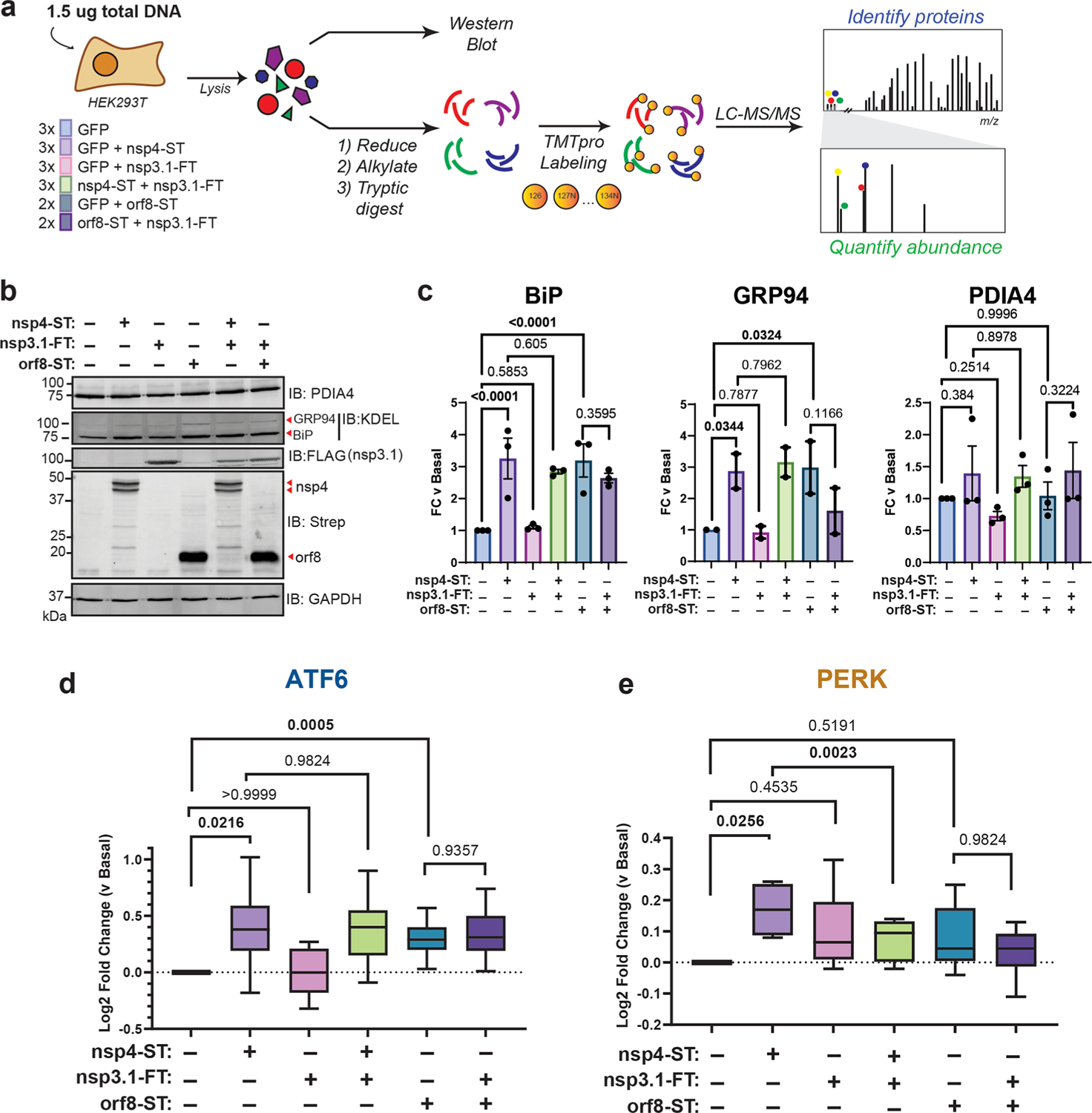
SARS-CoV-2 nsp3.1 suppresses nsp4-induced PERK, but not ATF6 activation. (a) Experimental schematic to test the effect of SARS-CoV-2 nsp3.1 on nsp4-or orf8-induced activation of the UPR in HEK293T cells by western blotting and tandem mass spectrometry (LC-MS/MS). (b) Representative western blot of ATF6 protein markers (PDIA4, BiP, GRP94) and viral proteins (SARS-CoV-2 nsp4-ST, nsp3.1-FT, and orf8-ST), with GAPDH as a housekeeping gene for a loading control. See Supplemental Figure S11 for all quantified, original blots. (c) Quantification of western blots in panel (b), normalized to GAPDH band intensities. n = 2–3, mean ± SEM, One-way ANOVA with Benjamini, Krieger, and Yekutieli multiple testing correction, p < 0.05 considered statistically significant. (d) Global proteomics analysis of ATF6 protein marker levels in the presence of SARS-CoV-2 nsp4-ST, nsp3.1-FT, orf8-ST, or specified combinations. Box-and-whisker plot shows median, 25th and 75th quartiles, and minimum and maximum values. One-way ANOVA with Geisser–Greenhouse correction and post hoc Tukey’s multiple comparison test was used to test significance; 1 MS run, n = 2–3 biological replicates. See Supplemental Tables S2 and S3 for mass spectrometry data set. (e) Global proteomics analysis of PERK protein marker levels, as in panel (d).
