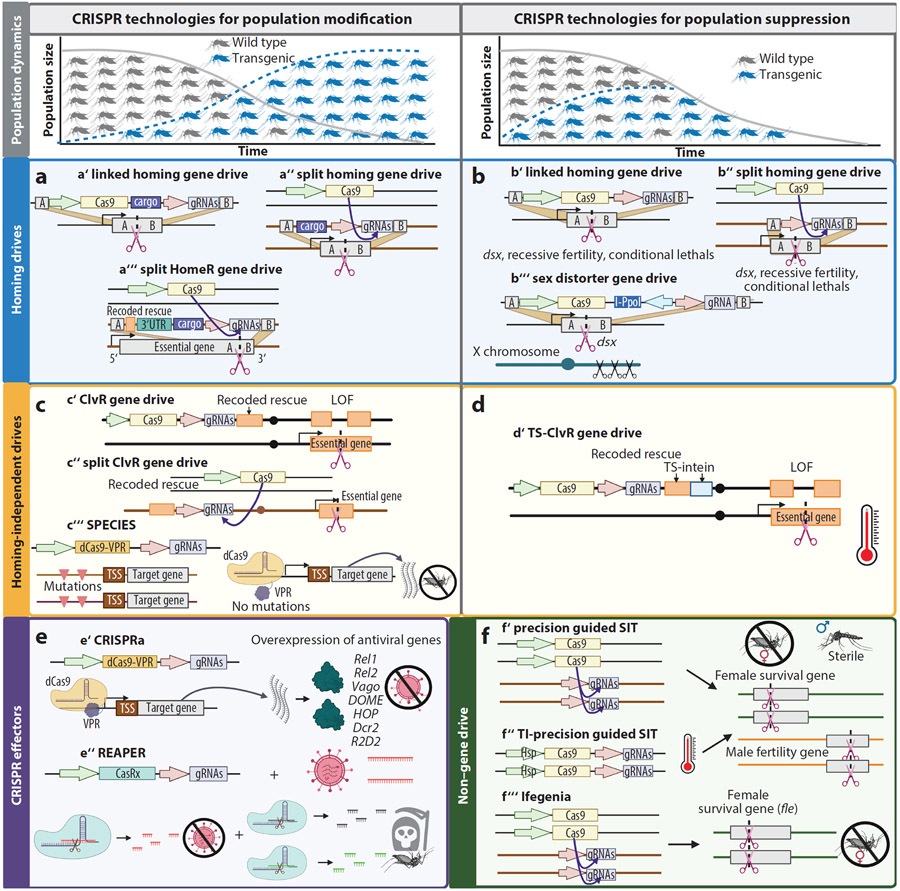
Figure 1.
Representative CRISPR technologies for the control of pest species. (Left panels) Population modification tools aim to drive desirable traits or cargo, such as disease resistance, into a population. (Right panels) Population suppression tools aim to reduce or eliminate the target population. As the transgene increases in frequency in the population, the population collapses. CRISPR technologies have been used to build (a,b) HCGDs, (c,d) homing-independent ClvR and (c) SPECIES, and (f) non–gene drive SIT technologies. (a) HCGDs for population modification typically target a unique, fitness-neutral location in the genome and then copy themselves and desirable effectors or cargo into the wild-type allele. The HomeR (a‴), for example, is a split drive designed to target an essential gene (the toxin), which is rescued by a linked cleavage-resistant version (the antidote) of the essential gene (124). HomeR was designed with a foreign 3′ UTR to prevent unintended recombination events, which can cause the generation of functional resistant alleles, and the drive was designed to target a 3′ locus in the essential gene, which minimizes the rescue recoding effort and uses the native promoter of the target gene to drive the expression of the rescue. (b) HCGDs for population suppression target either a recessive fertility gene, such as dsx, which creates sterile females when homozygous, or a conditionally lethal gene that can be driven to a high population frequency with minimal fitness load until subjected to a specific condition. HCGDs can be designed to either have both the Cas9 and gRNA gene drive elements linked at the same locus and inherited together (a′, b′) or have the Cas9 and gRNA unlinked at different loci (a″, b″) to reduce the frequency with which they are inherited together. When the Cas9 and gRNAs are unlinked, their reduced coinheritance makes the drive more spatially confinable than the linked drive, which has operational and safety considerations (see ethical considerations for low-threshold drives). Due to frequent unintended drive repair outcomes that build population resistance to a drive (see Figure 2), alternative designs are employed to reduce the formation of drive-resistant alleles in the population. For more stable and robust population suppression, the sex distorter HCGD developed by Simoni et al. (204), for example, homes into the recessive fertility gene dsx and includes a I-PpoI endonuclease to shred the X chromosome, generating infertile females when homozygous for the HCGD and biasing the population toward males (b‴). (c,d) Homing-independent methods have arisen in parallel to work on HCGDs. A TS ClvR drive (d′) contains a Cas9 and gRNA, which cleave a haplosufficient essential gene, and a linked rescue that is a recoded, cleavage-resistant version of the essential gene that also encodes a TS intein that disrupts the rescue at high temperatures (176). In this system, low temperatures allow the intein to splice out of the rescue gene for normal growth and drive fixation in the population, which is then disrupted at high temperatures when the intein is not spliced, so the essential gene is not rescued. Without this temperature specificity, ClvR can also be designed in a linked (c′) or split (c″) configuration to support population modification. The extent of component linkage can dictate the threshold, duration, and spread of the drive. The SPECIES system (c‴) uses a catalytically inactive version of Cas9 with a transactivator (dCas9-VPR) to cause untimely and lethal overexpression of an essential developmental gene. Lethality can be prevented by coinherited mutations that protect the target site from dCas9-VPR binding and lethal overexpression. (e) CRISPR-based effectors have been built to upregulate immunity genes (CRISPRa, e′) to decrease pathogen infection or to directly target and destroy RNA viruses and kill mosquitoes (REAPER, e″). When linked to a gene drive, these can replace a population that can transmit a pathogen with one that cannot sustain disease transmission. (f) pgSIT generates sterile males by simultaneously targeting genes required for male fertility and female survival early in embryogenesis (126, 128, 143). pgSIT uses the CRISPR Cas9 and gRNA components as either separate lines that generate sterile males only when crossed (f′) or a single line where the Cas9 expression is temperature inducible (TI-pgSIT, f″). In the TI-pgSIT system, Cas9 is expressed by an Hsp promoter and thus only at high temperatures (123). Ifegenia (f‴) is another binary non–gene drive CRISPR-based population control technology where separate Cas9 and gRNA lines are crossed to kill female offspring through the disruption of the fle gene (206). Here, male offspring harbor the female-killing mutation, which can lead to killing of any female offspring that result from their mating with wild females. Abbreviations: ClvR, Cleave and Rescue; CRISPRa, CRISPR activator; dsx, doublesex; fle, femaleless; gRNA, guide RNA; HCGD, homing CRISPR gene drive; HomeR, home and rescue toxin-antidote-based drive; Hsp, heat shock protein; ifegenia, inherited female elimination by genetically encoded nucleases interrupting alleles; LOF, loss of function; pgSIT, precision-guided sterile insect technique; REAPER, vRNA expression activates poisonous effector ribonuclease; SIT, sterile insect technique; SPECIES, synthetic postzygotic barriers exploiting CRISPR-based incompatibilities for engineering species; TI-pgSIT, temperature-inducible precision-guided sterile insect technique; TS, temperature sensitive; TSS, transcription start site; UTR, untranslated region; VPR, VP64-p65-Rta. Figure adapted from images created with BioRender.com.