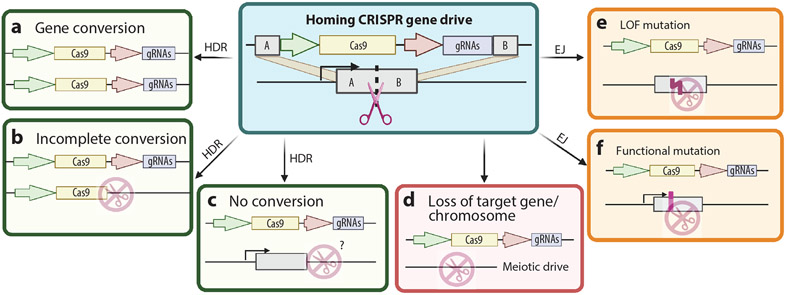
Figure 2.
HCGD outcomes. A simplified HCGD is shown using Cas9 and gRNAs to disrupt a gene via DSBs (teal box) that are repaired by HDR (green boxes) or EJ (orange boxes) but do not include cargo (e.g., resistant genes or recoded target gene). Alternatively, the repair process can fail altogether, resulting in the loss of a large chromosomal fragment (red boxes). (a) After cleavage of the target site, the HDR pathway uses the gene drive element as a template to repair the DSB, thereby copying the gene drive into the locus of the wild-type gene and biasing (increasing) the inheritance or copying of the gene drive. (b) Loss of parts of the HCGD can also occur during conversion when an alternative region of the gene drive element other than the homology arms is used as the template for the gene repair, such as part of the gRNA sequence, or as a result of internal recombination between the drive components. The chances of these events occurring increase with the size of the HCGD and multiplexing of gRNAs. Since the insertion site is still disrupted, such events would generate a drive-resistant allele that cannot be further cut by the HCGD. (c) No conversion (copying) may occur if the target site is not cleaved or the HDR process uses an alternate repair template, which could happen if the target locus has high homology to other regions in the genome; then the wild-type allele will be susceptible to cleavage and drive in subsequent generations. Otherwise, if the target site is repaired by a nonallelic template with high homology to the cut site it may be resistant to subsequent cleavage. (d) If a repair mechanism fails to repair the DSB, then the target gene, or in some cases the entire wild-type chromosome, may be lost. While this means the gRNA target site is no longer present, only gametes that inherit the drive will likely be viable since the wild-type gametes will be missing a chromosome or the target gene. This biased inheritance of gametes with the drive element is known as meiotic drive. (e,f) DSBs repaired by the EJ pathway can result in mutations at the target sites that make the next generation resistant to drive cleavage. (e) These mutations can cause a loss of gene function, which can cause the death of any offspring inheriting that mutation in a haploinsufficient gene. (f) If the mutation does not result in the loss of gene function, then the mutation inherited by the next generation will be resistant to drive cleavage. As these resistant alleles accumulate in a population over time, the drive will go extinct. Abbreviations: DSB, double-strand break; EJ, end joining; gRNA, guide RNA; HCGD, homing CRISPR gene drive; HDR, homology-directed repair; LOF, loss of function. Figure adapted from images created with BioRender.com.