FIGURE 3.
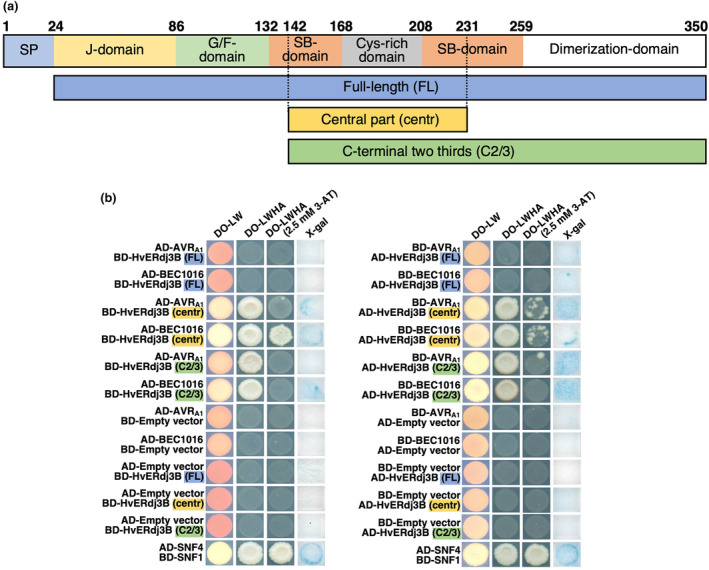
AVRA1 and BEC1016 interact with the central part of barley HvERdj3B in yeast two‐hybrid (Y2H) assay. (a) Domain structure of the 350 amino acid HvERdj3B protein and the Y2H fragments. The domains are predicted according to Chen et al. (2017) (see Figure S6). SP, signal peptide; G/F, glycine/phenylalanine; SB, substrate binding. Below the three Y2H fragments are shown. (b) Yeast transformed with the destination vectors pDEST‐AS2‐1 [GAL4 binding‐domain (BD)] and pDEST‐ACT2‐1 [GAL4 activation‐domain (AD)] (Robertson, 2004) in different combinations of domain fusions to effectors and the full‐length (FL), the central part (centre) and the C‐terminal two‐thirds (C2/3) of HvERdj3B. Growth on dropout (DO) medium lacking leucine (L) and tryptophan (W) indicated presence of both constructs. Growth on DO medium lacking L, W, histidine (H) and adenine (A) ± 2.5 mM 3‐amino‐1,2,4‐triazole (3AT) indicated protein–protein interaction. β‐galactosidase assay (X‐gal), also indicating protein–protein interaction, was performed on filter paper prints of DO‐LW‐grown yeast. SNF1/SNF4 was used as a positive control and empty vectors were used as negative control.
