Fig. 4 |. NG PAM Cas9 base editors enable high-resolution screens.
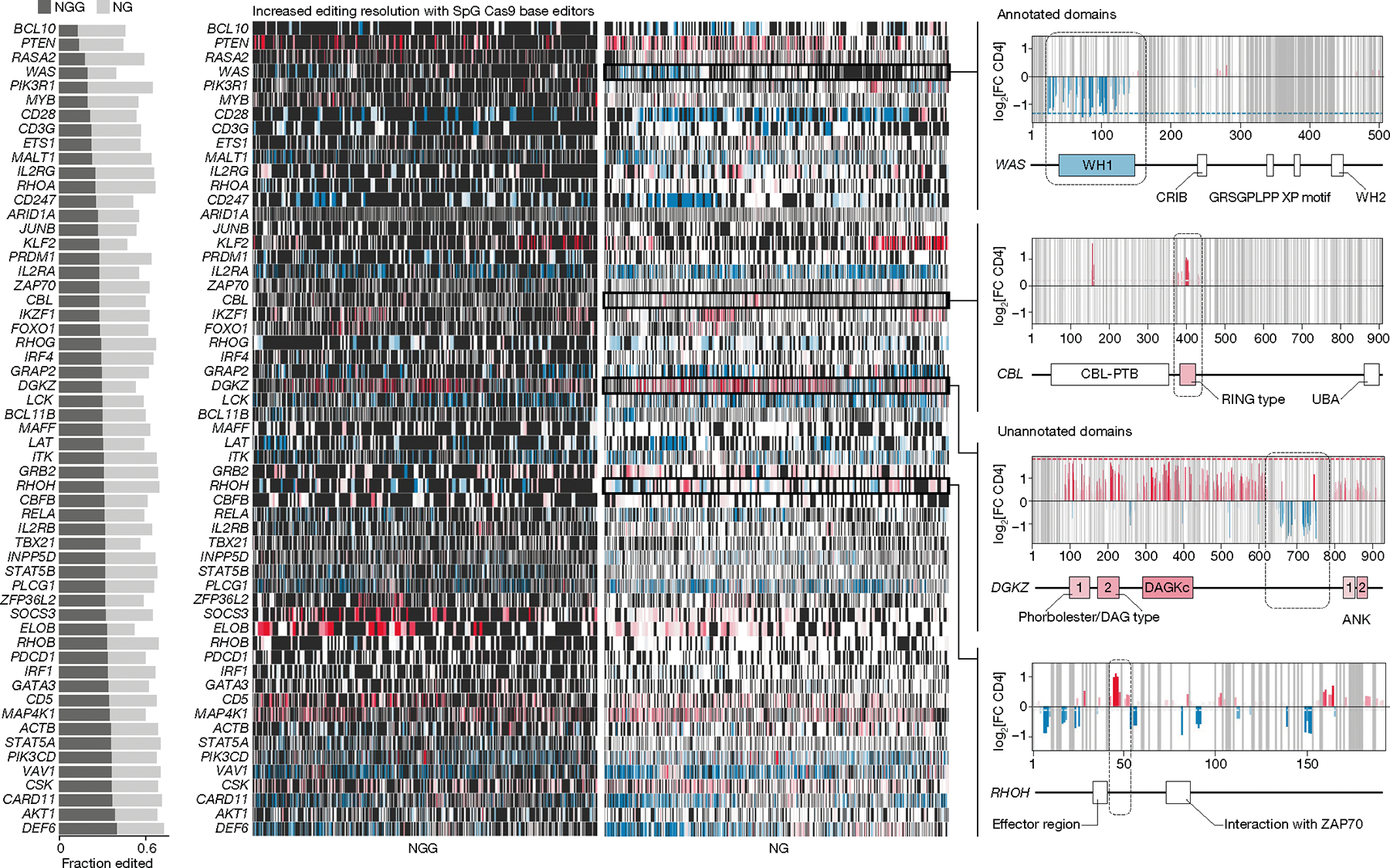
Comparison of base-editing resolution between NGG PAM- and NG PAM-dependent Cas9 for 57 genes overlapping between libraries. Left, the fraction of editable residues with NGG PAM compared with NG PAM ABE base editors. Centre, heat maps of effects of base editing across coding sequences for each gene (rows); black indicates an absence of data (uneditable residue), white is an editable residue with no observed effect, red indicates a positive effect, and blue indicates a negative effect. Right, detailed results for base edits across WAS, CBL, DGKZ and RHOH, with domains confirmed by base editing (WAS and CBL) or suggested as novel functional domains (DGKZ and RHOH) boxed and indicated on the gene plots. X-axis labels on far right plots show amino acid positions. CRIB, Cdc42- and Rac-interactive binding motif; DAGKc, diacylglycerol kinase catalytic domain; PTB, phosphotyrosine-binding domain; WH, WASp homology domains.
