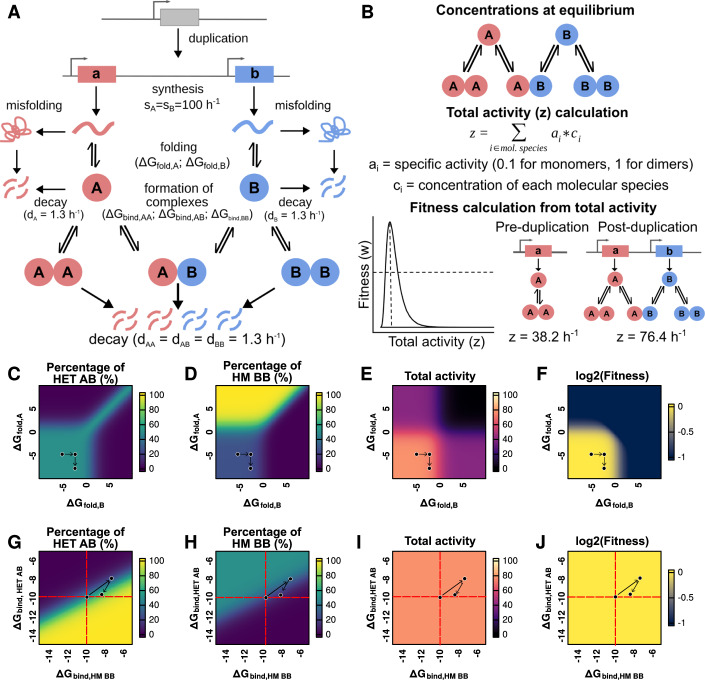
Figure 1. Changes to folding free energy and binding affinity modify the concentration of each dimer and the fitness of the system.
(A) Post-duplication system. Paralogous proteins are produced with synthesis rates sA and sB. Synthesis encompasses both transcription and translation, which are not considered separately. The probability of correct folding is calculated based on the folding free energy (ΔGfold) (see “Methods”). Misfolded proteins are removed from the system. Folded proteins can assemble into homo- or heterodimers based on binding free energies (ΔGbind,HM AA, ΔGbind,HET AB, ΔGbind,HM BB). (B) The concentrations of each molecular species (monomers, homodimers, and heterodimers) are estimated at equilibrium. Total protein activity is calculated by multiplying the equilibrium concentrations by their specific activity (0.1 for monomers, 1 for both homodimers and the heterodimer). Fitness is estimated based on the total protein activity using a lognormal function given by parameters alpha and beta. With the parameters for synthesis rates and decay rates shown in (A), the estimated values of total activity for the system before and after the duplication corresponded to 38.2 h−1 and 76.4 h−1. (C–F) Effect of variation in the ΔGfold for each monomer on the percentage of heterodimer (HET AB) (C), percentage of one of the homodimers (HM BB) (D), the total activity (E), and fitness (F) of the system. Fitness is estimated using a lognormal distribution with the optimum (alpha value) set to a total activity of 80 (see (B)). Arrows represent an example of a mutational trajectory. (G–J) Effect of variation in ΔGbind,HET AB and ΔGbind,HM BB on the percentage of the heterodimer (HET AB) (G), percentage of one of the homodimers (HM BB) (H), and the total activity (I), and fitness (J) of the system. (G–J) ΔGbind,HM AA was kept constant at −10 kcal /mol, indicated by the red dashed lines. Arrows represent an example of a mutational trajectory.