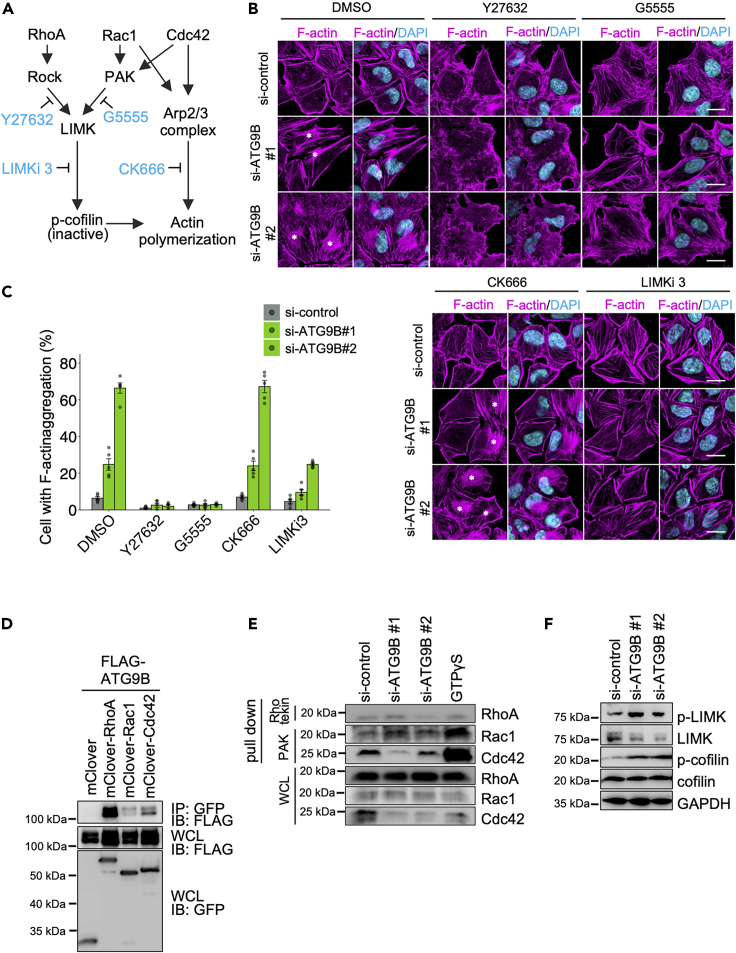
Figure 4.
ATG9B modulates actin rearrangement in HeLa cells
(A) Schematic representation of the RhoGTPase pathway and the pathways blocked by the respective inhibitors.
(B and C) Treatment of ATG9B siRNA-treated HeLa cells with ROCK inhibitor (Y27632), PAK inhibitor (G5555), LIMK inhibitor LIMKi 3, or Arp2/3 complex inhibitor (CK666) for 2 h. Cells were fixed and immunostained for F-actin (magenta), and cellular DNA was stained with DAPI (cyan). Asterisks indicate actin aggregation. Representative microscopic images (B) and quantification of cell percentage with F-actin aggregation (C).
(D) Co-IP between FLAG-tagged ATG9B and mClover-tagged Rho GTPase proteins.
(E) Immunoblotting of Rho-binding domain or p21-binding domain agarose-isolated active RhoA, Rac1, or Cdc42 from whole cell lysates (WCL) obtained from ATG9B siRNA-treated HeLa cells.
(F) Immunoblotting analysis of ATG9B siRNA-treated HeLa cells with the indicated antibodies.Data in c (n = 100 cells per condition) represent the mean ± SEM from five independent experiments: scale bar, 10 μm. One-way ANOVA was performed and p values were calculated using Tukey’s multiple comparison test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.