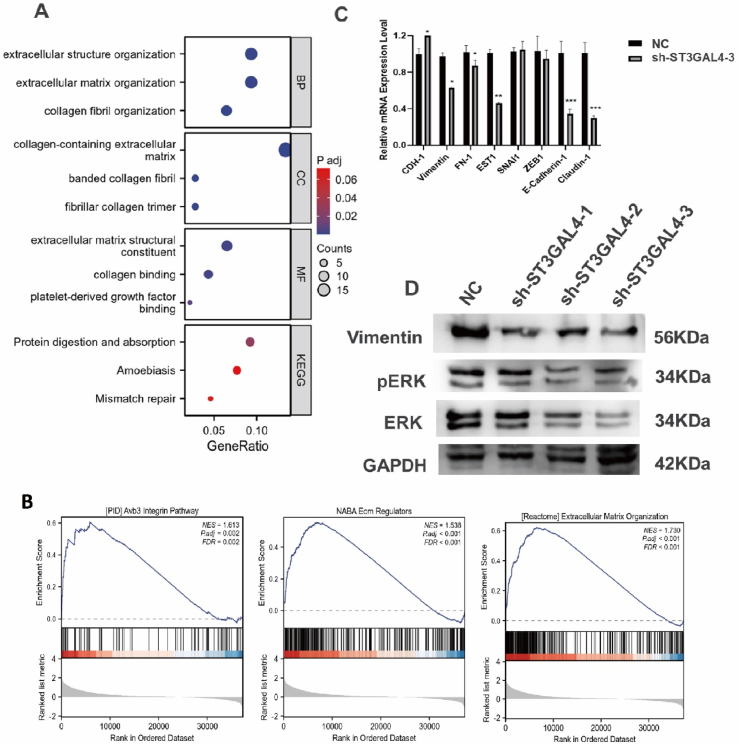
Fig. 5.
Functional enrichment analysis demonstrated the involvement of ST3GAL4 in extracellular matrix organization. A: Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed on the top 200 upregulated differentially expressed genes (DEGs) in glioma patients with high ST3GAL4 expression. B: The gene set enrichment analysis (GSEA) identified pathways related to the extracellular matrix that were significantly enriched. C: qRT-PCR analysis to evaluate the expression of eight representative epithelial-mesenchymal transition (EMT) biomarkers in U251 cells with or without ST3GAL4 knockdown. *P < 0.05, **P < 0.01, ***P < 0.001. D: Western blot analysis to examine the protein expression levels of vimentin, ERK1/2, pERK1/2, and GAPDH in U251 cells.