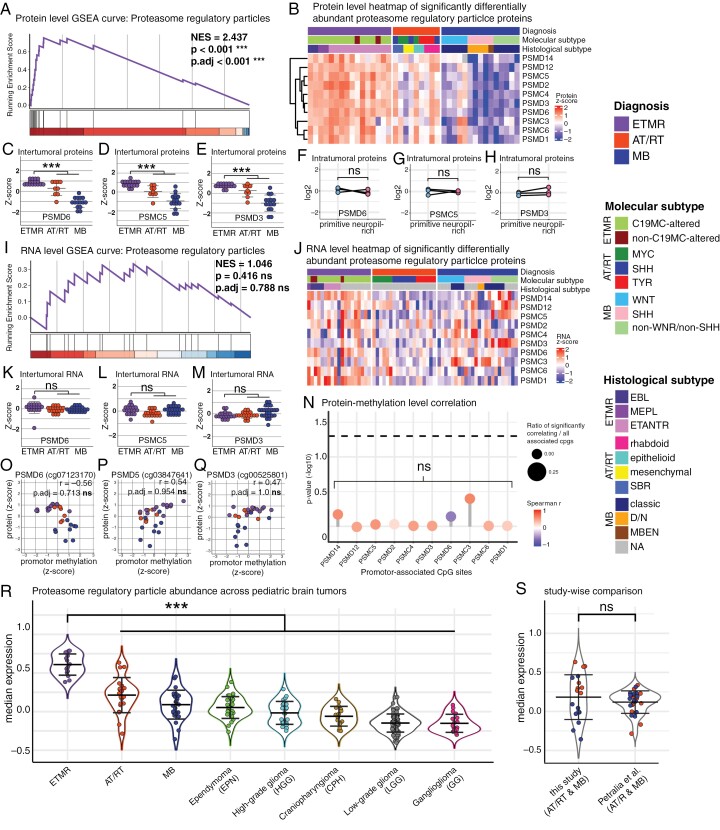
Figure 5.
Proteasome regulatory particle proteins are highly abundant in ETMR (A) GSEA curve of the “proteasome regulatory particle” gene set based on protein abundancies comparing ETMR to AT/RT and MB. P-value was BH-adjusted. (B) Proteome heatmap shows a pervasive abundance of proteasome regulatory particles in ETMR compared to AT/RT and MB. (C—E) Plots show protein levels of the 3 top ranked proteasome regulatory particle gene set members PSMD6 (C), PSMC5 (D), and PSMD3 (E). ***P < .001. Welch’s t-test. (F—H) Microdissection analyses confirm that protein abundance of PSMD6 (F), PSMC5 (G), and PSMD3 (H) is not associated with histomorphology of ETMR. (I) GSEA curve of the “proteasome regulatory particle” gene set based on RNA levels comparing ETMR to AT/RT and MB shows no significant enrichment for ETMR. P-value was BH-adjusted. (J) Transcriptome heatmap of “proteasome regulatory particle” RNA levels appears noisy across embryonal brain tumors. (K—M) Plots show RNA levels of the 3 top ranked proteasome regulatory particle gene set members PSMD6 (K), PSMC5 (L), and PSMD3 (M). No significant fold changes can be seen. P-values were BH-adjusted. ns: not significant. Welch’s t-test. (N) Lollipop plot shows the correlation of protein and promotor methylation levels of “proteasome regulatory particle” gene set members. No members show a significant correlation with any CpG sites associated with their respective promotor regions. Spearman correlation. P-values were BY-adjusted. ns = not significant. (O—Q) Plots show protein levels and the methylation levels of the representative promotor CpG sites across embryonal brain tumors. No significant correlation is seen in the 3 top ranked proteasome regulatory particle gene set members PSMD6 (O), PSMC5 (P), and PSMD3 (Q). (R, S) Integrated proteome data of our study and publicly available proteome data of further pediatric brain tumors (Petralia et al.) demonstrates that the abundancy of proteasome regulatory particles is a prominent molecular feature of ETMR. Median protein abundance of all “proteasome regulatory particle” gene set members are shown across entities (R) and comparing overlapping entities (AT/RT and MB) represented in both datasets (S). The latter plot confirms that differential protein levels do not relate to batch effects. ***P < .001. Welch’s test.