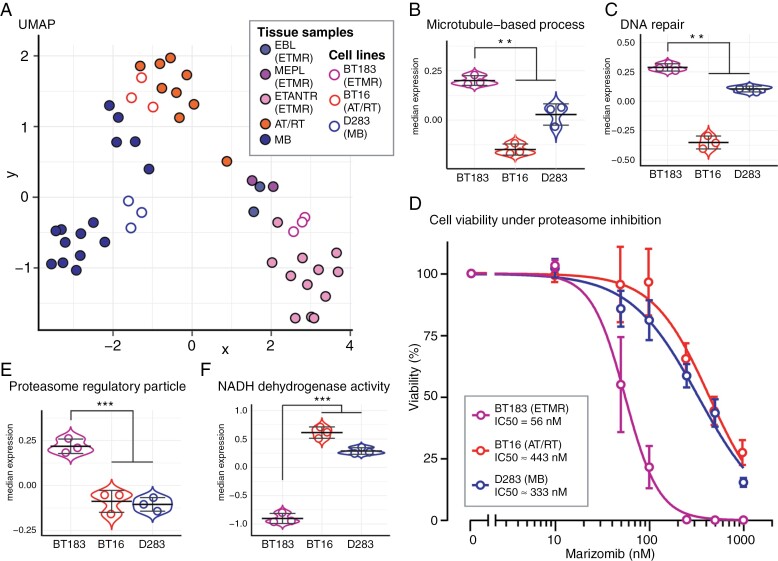
Figure 6.
Proteasome inhibition represents a promising therapeutic vulnerability in ETMR (A) UMAP plot shows that the proteome profiles of the cell lines BT183, BT16, and D283 adequately match with ETMR, AT/RT, and MB tumor tissue, respectively. UMAP dimension reduction was based on the top 1000 most significant proteins of an ANOVA test between ETMR, AT/RT and MB tissue samples. (B—C) Median protein abundance of all “Microtubule-based process” and “DNA repair” gene set members are shown in BT183, BT16 and D283. Proteins of both gene sets are significantly more abundant in BT183 compared to BT16 and D283. **P < .01. Welch’s t-test. (D) Cell titer glo viability assay of BT183 (ETMR), BT16 (AT/RT), and D283 (MB) cell lines treated with the CNS penetrant proteasome inhibitor Marizomib at different concentrations. BT183 ETMR cells are highly vulnerable towards Marizomib treatment, also in comparison with other embryonal tumor cell lines. Four parameter logistic (4PL) curve regression model. (E—F) Median protein abundance of all “Proteasome regulatory particles” and “NADH dehydrogenase activity” gene set members are shown in BT183, BT16, and D283. Proteasome regulatory particle proteins are highly abundant in BT183 compared to BT16 and D283. In contrast, NADH dehydrogenase activity proteins are significantly less abundant in BT183 compared to BT16 and D283. ***P < 0.001. Welch’s t-test.