FIG. 1.
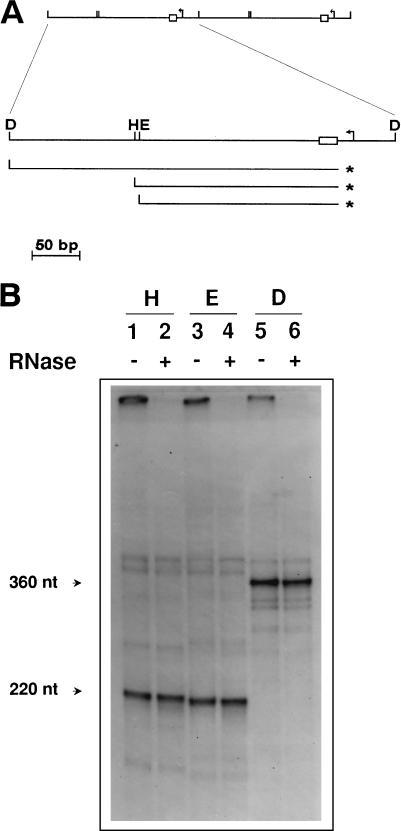
Detection and mapping of free 5′ ends in the mtDNA of strain a-1/1R/Z1. (A) Schematic diagram of the repeat units of the ori1 petite mutant showing the results of the 5′ mapping experiment illustrated in panel B. The upper part shows two repeat units of the mitochondrial genome of a-1/1R/Z1. Below is an enlargement of one such repeat showing the promoter (bent arrow) and the GC cluster C element (box) present in the ori1 sequence, as well as the positions of the DraI (D), HpaII (H), and EcoRV (E) restriction sites used in the analysis. Also indicated are the position of the predominant 5′ end detected (asterisks) and the strands containing this terminus upon restriction enzyme digestion and denaturation. (B) Detection of free 5′ ends by direct labeling with [γ-32P]ATP. mtDNA from a-1/1R/Z1 was dephosphorylated and labeled with [γ-32P]ATP prior to restriction enzyme digestion and electrophoresis on a denaturing polyacrylamide gel. Lanes were pretreated with RNases as indicated (+). The restriction enzymes used were HpaII (lanes 1 and 2), EcoRV (lanes 3 and 4), and DraI (lanes 5 and 6).