Abstract
mRNA translation and decay are tightly interconnected processes both in the context of mRNA quality-control pathways and for the degradation of functional mRNAs. Cotranslational mRNA degradation through codon usage, ribosome collisions, and the recruitment of specific proteins to ribosomes is an important determinant of mRNA turnover. However, the extent to which translation-dependent mRNA decay (TDD) and translation-independent mRNA decay (TID) pathways participate in the degradation of mRNAs has not been studied yet. Here we describe a comprehensive analysis of basal and signal-induced TDD and TID in mouse primary CD4+ T cells. Our results indicate that most cellular transcripts are decayed to some extent in a translation-dependent manner. Our analysis further identifies the length of untranslated regions, the density of ribosomes, and GC3 content as important determinants of TDD magnitude. Consistently, all transcripts that undergo changes in ribosome density within their coding sequence upon T cell activation display a corresponding change in their TDD level. Moreover, we reveal a dynamic modulation in the relationship between GC3 content and TDD upon T cell activation, with a reversal in the impact of GC3- and AU3-rich codons. Altogether, our data show a strong and dynamic interconnection between mRNA translation and decay in mammalian primary cells.
mRNA degradation contributes to the definition of steady-state transcript levels, removing aberrant mRNAs through surveillance pathways, as well as to the dynamic regulation of mRNA abundance in response to cellular cues. mRNA degradation involves a wide variety of effectors and regulatory factors, which for the most part correspond to RNA-binding proteins that are able to recognize specific sequences or structural motifs in the target mRNAs. Similar to most other steps in the gene expression pathway, mRNA degradation is often coordinated with upstream and downstream steps. For example, mRNA translation and decay are strongly interconnected in the context of mRNA surveillance pathways (Bicknell and Ricci 2017; Morris et al. 2021). Translation-dependent decay (TDD) pathways were originally identified and studied as the means by which cells rid themselves of aberrant mRNAs (e.g., those containing premature termination codons in the case of nonsense mediated decay [NMD] and truncated or prematurely polyadenylated mRNAs in the case of nonstop decay [NSD]). However, it is now widely recognized that NMD is also a key post-transcriptional regulatory mechanism for physiologically functional mRNAs (Nasif et al. 2018). Furthermore, other TDD pathways have been discovered that regulate the stability of physiologically functional transcripts bearing binding sites for specific RNA-binding proteins (Kim et al. 2005; Mino et al. 2015; Hia et al. 2019), through specific codon usage (Presnyak et al. 2015; Bazzini et al. 2016; Mishima and Tomari 2016; Radhakrishnan et al. 2016; Hanson and Coller 2018; Hia et al. 2019; Narula et al. 2019; Wu et al. 2019; Forrest et al. 2020; Medina-Muñoz et al. 2021), through the nascent peptide amino acid composition (Forrest et al. 2020; Burke et al. 2022), or as a consequence of ribosome collisions (Garzia et al. 2017; Sundaramoorthy et al. 2017; Juszkiewicz et al. 2018; Tuck et al. 2020). Cotranslational mRNA degradation (Hu et al. 2009; Pelechano et al. 2015; Ibrahim et al. 2018; Wu et al. 2019; Tuck et al. 2020; Medina-Muñoz et al. 2021; Bae and Coller 2022; Dave et al. 2023) has therefore emerged as a major decay pathway for functional mRNAs in eukaryotic cells. However, the extent to which TDD is a general mechanism for limiting the number of protein molecules made per mRNA molecule, both basally and in response to signaling, is currently unknown. Furthermore, the relationship between TDD and translation-independent mRNA decay (TID) pathways has not yet been evaluated in a global manner.
Here we describe a comprehensive analysis of basal and signal-dependent gene expression in primary resting and activated mouse CD4+ T lymphocytes. These cells represent an interesting model as they transition from quiescence to a proliferative state that requires profound changes in their gene expression profile and metabolism (Nicolet et al. 2021). Here, using a combination of transcription and translation inhibitors together with RNA-seq, ribosome profiling, and poly(A)-site sequencing, we aimed at discriminating between TDD and TID in a transcriptome-wide manner and identify features associated with each degradation pathway in both resting and activated cells.
Results
Inhibition of translation stabilizes numerous transcripts in T cells
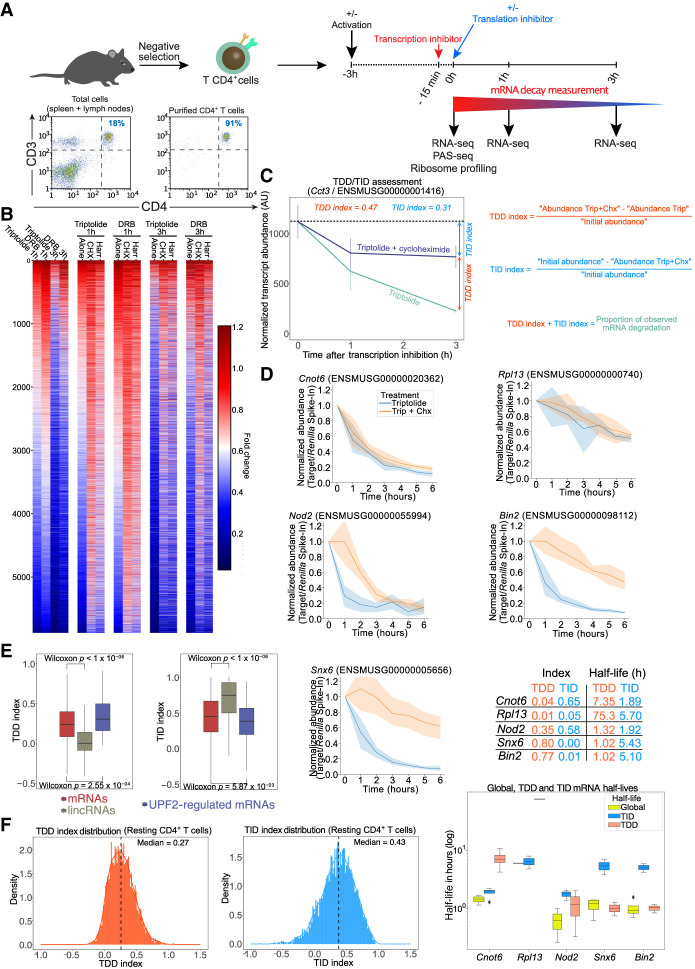
Purified primary mouse CD4+ T lymphocytes (>90% CD3+ CD4+) (Fig. 1A) were obtained by negative selection from C57BL/6J mouse spleens and lymph nodes. To monitor mRNA decay rates in the presence and absence of translation, we used a transcription inhibition strategy (Fig. 1A) in order to avoid any potential bias introduced by a possible coupling between translation and RNA transcription (Timmers and Tora 2018). For this, we tested the efficiency of two different transcription inhibitors, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), which inhibits RNA polymerase II elongation (Dubois et al. 1994), and triptolide, which irreversibly blocks transcription from RNA polymerase II through targeting of the general transcription factor TFIIH (Titov et al. 2011). We did not use metabolic labeling with 4-thiouridine to monitor mRNA stability because overnight incubation of cells with the modified nucleotide led to cell toxicity (Supplemental Fig. S1) and has been shown to inhibit rRNA synthesis, causing nucleolar stress (Burger et al. 2013). Cells exposed to each of the transcription inhibitors were collected after 1 and 3 h, and total RNA was extracted to prepare the RNA-seq libraries (Fig. 1A; Supplemental Fig. S2A). Comparison of the degradation fold observed at 3 h for both drugs indicated an overall good correlation (r = 0.77, Pearson), although it skewed in favor of increased observed degradation for the triptolide condition (Supplemental Fig. S2B). This difference could be owing to a difference in the speed at which each inhibitor effectively blocks transcription.
Figure 1.
Monitoring translation-dependent and -independent mRNA decay in CD4+ T lymphocytes. (A) Schematic representation of the procedure to monitor overall and translation-dependent mRNA degradation. Purified CD4+ T lymphocytes, as shown by flow cytometry analysis of CD3 and CD4 surface expression in cells obtained from spleen and lymph nodes before and after purification by negative selection, are incubated with transcription inhibitors (triptolide or DRB). Fifteen minutes following addition of transcription inhibitors, translation inhibitors (cycloheximide or harringtonine) are added to cells. Cells are collected at 0, 1, and 3 h following transcription inhibition to monitor transcript expression by RNA-seq. Ribosome profiling and poly(A)-site sequence (PAS-seq) were also performed at the time 0 h in absence of transcription inhibitors. (B) Fold-change in transcript abundance (compared with the 0-h time point) upon blocking transcription only or when blocking both transcription and translation with different drug combinations. (C) Details of the calculation of the TDD and TID indexes illustrated by the expression dynamics of the transcript coding for CCT3 (ENSMUSG00000001416) upon incubation with transcription and translation inhibitors. (D) mRNA half-life measurement of TDD and TID targets by quantitative PCR in resting T cells incubated with triptolide or triptolide + cycloheximide. (E) Comparison of the TDDindex (left) or TIDindex (right), obtained using triptolide and cycloheximide, for protein-coding transcripts (mRNAs), long intergenic noncoding RNAs (lincRNAs), and UPF2-regulated transcripts in resting T cells. Displayed P-values correspond to the mean P-value of a Wilcoxon test performed 1000 independent times on samples of the same size for each compared group. (F) Distribution of the TDDindex and TIDindex calculated from resting T cells incubated for 3 h in the presence of triptolide and cycloheximide.
To assess the extent to which mRNA decay is dependent on translation, we inhibited translation using cycloheximide, which blocks elongating ribosomes on the coding sequence, or using harringtonine, which blocks the late steps of translation initiation, resulting in the run-off of elongating ribosomes and the accumulation of initiating 80S ribosomes at translation start sites (Fig. 1A; Supplemental Fig. S2C). Briefly, T cells were incubated with the above-mentioned combinations of inhibitors for different time points (0 h, 1 h, or 3 h after transcription or transcription plus translation inhibition), and RNA levels were assessed by RNA-seq from three independent biological replicates for all conditions.
Results obtained using either DRB or triptolide showed that translation inhibition, whether through cycloheximide or harringtonine, led to a global stabilization of mRNAs at both the 1-h and 3-h time points (Fig. 1B). Furthermore, stabilization was observed across the entire length of transcripts (including untranslated regions [UTRs]) in both cycloheximide- and harringtonine-treated cells (Supplemental Fig. S2D–F), with no detectable increase in reads in the coding sequence observed after blocking translation. This indicates that the observed stabilization does not result from the physical presence of ribosomes in the coding sequence that could block the progression of exonucleases and lead to a bias toward reads in the coding sequence.
To quantify the extent to which overall mRNA decay is dependent on ongoing translation, we calculated a translation-dependent RNA decay index for each time point (TDDindex) (Fig. 1C). This index corresponds to the difference in absolute transcript abundance between the blockade of both transcription and translation and the blockade of transcription only, normalized by the initial transcript abundance before transcription inhibition (for TDDindex formula, see Fig. 1C). Similarly, we calculated a translation-independent RNA decay index (TIDindex), which measures the difference between transcript abundance at the initial time and after blocking of transcription and translation, normalized by the initial transcript abundance (Fig. 1C, see TIDindex formula). For any given transcript, the sum of the TDDindex and TIDindex corresponds to the fraction of mRNAs that have been degraded at any given time after blocking transcription (Fig. 1C). Transcripts whose decay is largely translation independent have TDDindex values close to zero and TIDindex values greater than zero, whereas transcripts whose decay is mainly translation dependent have a TDDindex greater than zero and TIDindex close to zero. Because mRNA degradation generally follows exponential decay kinetics (although it is not the case for all mRNAs, particularly in mammalian cells, which have complex multistep degradation pathways), the TDD and TID indexes should only provide semiquantitative information regarding the extent of TDD and TID. To validate that TDD and TID indexes provide semiquantitative information regarding the extent of TDD and TID on mRNA half-lives, we monitored mRNA half-lives for five different transcripts during a 6-h time course in the presence of triptolide or triptolide + cycloheximide (Fig. 1D). We chose one transcript mainly degraded through TID (Cnot6), one relatively stable transcript mainly degraded through TID (Rpl13), one transcript degraded both through TDD and TID (Nod2), and two transcripts mainly degraded through TDD (Snx6 and Bin2). Observed mRNA half-lives are in agreement with calculated TDD and TIDindex (Fig. 1D), indicating that TDD and TID indexes can be used as a proxy to monitor each degradation pathway.
TDDindexes obtained from the different combinations of transcription and translation inhibitors yielded very similar values, with Pearson correlation coefficients ranging from 0.67 to 0.82 (Supplemental Fig. S3A,B), indicating a robust measure of TDD independently of the transcription or translation inhibitors that are used. We nevertheless observe a general skewing of the population toward higher TDD index values for the cycloheximide conditions compared with the harringtonine-treated cells. This difference could be because harringtonine allows elongating ribosomes to run-off during several minutes after drug addition, possibly further destabilizing the mRNA in a translation-dependent manner during that time. Finally, to rule out any technical bias introduced by the use of transcription inhibitors, we performed RNA-seq measurements in cells incubated for 3 h in the presence, or not, of either cycloheximide or harringtonine alone (in the absence of transcription inhibitors). From these data sets, we calculated the fold-change in transcript abundance upon blocking translation and used it as a proxy for the TDDindex. As shown (Supplemental Fig. S3C,D), TDDindexes calculated using transcription and translation inhibitors correlate with the fold-change in transcript abundance obtained upon translation inhibition for both cycloheximide and harringtonine, although to a lesser extent than the correlations observed when comparing TDDindexes obtained with different combinations of transcription and translation inhibitors. These differences can be explained by possible feedback regulatory loops that could modulate transcription in response to changes in mRNA translation and degradation rates as these processes can be coupled within cells (Timmers and Tora 2018; Slobodin and Dikstein 2020).
To validate our experimental strategy to monitor TDD, we examined the extent of translation-dependent RNA decay of long intergenic noncoding RNAs (lincRNAs). lincRNAs are transcribed by RNA polymerase II and share many features with protein-coding mRNAs, including a 5′ cap structure, excision of introns by the spliceosome, and a 3′ poly(A) tail (Zhang et al. 2016). However, they are mostly not engaged with the translational machinery and therefore should not be decayed in a translation-dependent manner. Consistent with this, the TDDindex of lincRNAs is centered around zero (Fig. 1E, left), whereas their TIDindex is close to 0.75, a value significantly higher than that of protein-coding mRNAs (Fig. 1E, right). Therefore, as expected, lincRNAs are mainly degraded through a translation-independent pathway. Conversely, the median TDDindex of transcripts previously identified in mouse thymocytes as regulated by UPF2 through NMD, a TDD pathway (Weischenfeldt et al. 2008), is significantly higher than the median TDDindex of all protein-coding mRNAs (Fig. 1E, left), whereas their TIDindex is significantly lower (Fig. 1E, right). These results show that our calculated TDD and TID indexes behave as expected for transcripts whose decay should be either independent (lincRNAs) or dependent (NMD targets) on translation.
Distribution of the TDDindex calculated at 3 h upon blocking transcription (Fig. 1F, left) shows a median value of 0.27 (i.e., half of all the expressed transcript species display a decrease in initial abundance of at least 27% that is dependent on translation), thus confirming our previous observation that a large fraction of the transcripts are degraded, at least partially, in a translation-dependent manner. A small fraction of transcripts displayed high TDDindex values (close to one), corresponding to highly unstable mRNAs relying almost entirely on translation for their decay (Fig. 1F, left). Transcripts with a high TDDindex are enriched in Gene Ontology (GO) categories related to core gene expression functions such as ribosome biogenesis, mRNA splicing, tRNA processing, RNA helicase activity, and the proteasome complex (Supplemental Fig. S4A). GO categories such as DNA repair and mitochondrial inner membrane were also enriched in transcripts with a high TDDindex. A larger fraction of transcripts displayed TDDindex values close to zero, indicating that their degradation is not dependent on translation (Fig. 1F, left). These transcripts are enriched in GO categories related to RNA pol II transcription factors, proteins associated to the plasma membrane, signal transduction, protein kinase activity, and ribosomal proteins (Supplemental Fig. S4A).
Distribution of the TIDindex is overall broader than that of the TDDindex (Fig. 1F, right) and displays a higher median value of 0.43 (i.e., half of all the expressed transcript species display a decrease in initial abundance of at least 43% that is independent of translation). Overall, TID is preponderant over TDD in defining the extent of mRNA decay in T cells (Supplemental Fig. S2G). Some functional categories enriched in low and high TID transcripts showed a mirror image of those observed for TDD (Supplemental Fig. S4A,B). For example, transcripts coding for RNA pol II transcription factors are enriched among high TIDindex and low TDDindex groups. In contrast, transcripts coding for DNA repair and mitochondrial membrane proteins are enriched among low TIDindex and high TDDindex groups. Transcripts encoding RNA splicing factors appear strongly regulated by both TDD and TID. Conversely, other categories such as mRNAs coding for ribosomal proteins, synapses, or protein serine/threonine kinases are enriched in both low TDD and TID transcript groups, therefore indicating that they correspond to stable transcripts. Finally, GO categories related to the immune response (such as innate immune response, cellular response to lipopolysaccharide, and immune system process) appear to be mostly regulated through a TID pathway.
In conclusion, our data indicate that TDD is involved in the degradation of a significant fraction of cellular mRNA species, although to a lesser extent than TID, which is the main pathway of mRNA degradation in T cells.
mRNA features defining the extent of TDD in T CD4+ lymphocytes
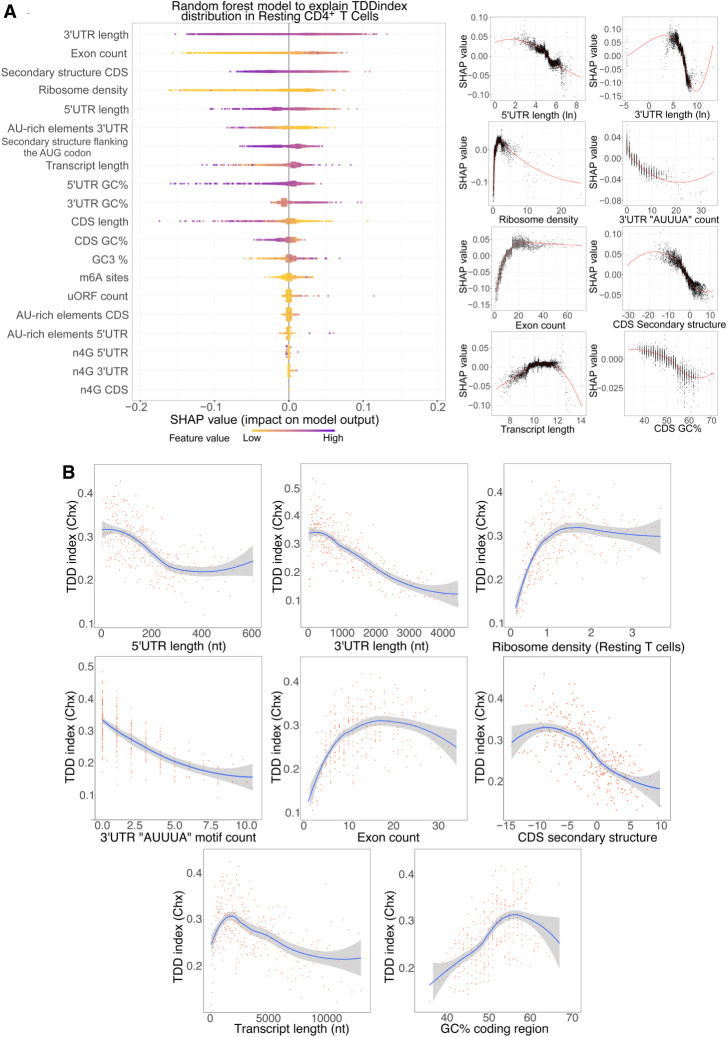
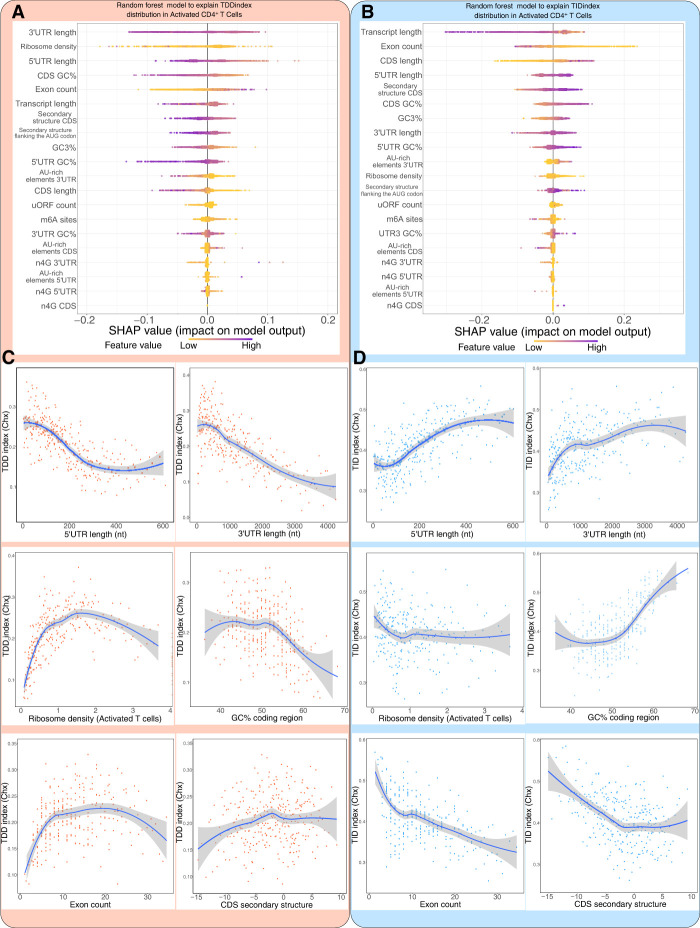
Having uncovered a global impact of mRNA translation on mRNA decay in T cells, we next investigated which mRNA features could explain the extent of TDD. To this aim, we built a random forest model (Supplemental Code S1) to predict the observed TDDindex based on transcript-related information such as total transcript length, length of the 5′ UTR, coding sequence and 3′ UTR length (obtained from our PAS-seq data performed in resting and activated T CD4+ cells), ribosome density at the coding sequence (obtained from our ribosome profiling data performed in resting and activated T CD4+ cells), density of m6A sites (experimentally obtained from T CD4+ mouse cells) (Li et al. 2017), or the number of upstream open reading frames (uORFs), among others. The model was trained with 80% of our data set, whereas the remaining 20% was used as a test set. In the test set, the trained model was able to capture 32% of the observed variance (to visualize predicted versus observed TDDindexes in the training and validation data sets, see Supplemental Fig. S5A). This result suggests that additional transcript features, such as information regarding RNA binding proteins or microRNA binding sites, which were not included in our data set, could participate in mediating TDD in addition to those tested in our study. To interpret the model predictions and visualize the quantitative contribution of each feature to the model's prediction, we used shapley additive explanation (SHAP) values (Fig. 2A; Rodríguez-Pérez and Bajorath 2020).
Figure 2.
Characterization of cis- and trans-acting features linked to TDD. (A) Random forest decision tree analysis of transcript features to explain the observed TDDindex values in resting CD4+ T lymphocytes coupled with SHAP value analysis for interpreting the output of the random forest model. (Left) Features are sorted from top to bottom with respect to their importance in predicting the TDDindex and SHAP values displayed to show how much each feature has influenced the model. (Right) Plots showing the impact of SHAP values on the model for the main identified features. (B) Binning plots of the TDDindex against selected features. Transcripts are first ordered with respect to the feature to be compared with the TDDindex and groups of 20 transcripts made along the selected feature. The mean TDDindex and feature values are plotted for each group.
Analysis of the SHAP values from the random forest model led to the identification of 3′ UTR length, ribosome density, and 5′ UTR length among the main features predicting the observed TDDindex (Fig. 2A). These results are consistent with a direct role of ribosomes in mediating decay of the mRNA they translate. SHAP value plots of 5′ UTR and 3′ UTR length show a clear negative correlation between these two transcripts features and their corresponding SHAP values (Fig. 2A, right), indicating that transcripts with short 3′ UTR or short 5′ UTR are more prone to TDD than those with long UTRs (Fig. 2B). To validate this observation, the relationship between these factors (5′ UTR and 3′ UTR length) and the observed TDDindex was plotted using binned data (Fig. 2B). For this, transcripts were ordered based on the variable of interest (5′ UTR length or 3′ UTR length), and bins of transcripts (each bin containing 20 transcripts with consecutive values for the variable of interest) were made. Then, the mean TDDindex and variable of interest values are plotted for each bin. This binning strategy allows better visualization of the relationship between the variable of interest and the TDDindex by smoothing the contribution of the other variables that affect TDD.
Ribosome density shows a monotonic with saturation relationship with the TDDindex, displaying a strong positive correlation for low-to-medium ribosome density values that reaches a plateau for high ribosome densities (Fig. 2B). This is confirmed by the SHAP (Fig. 2A, right). Transcripts bearing low ribosome densities are therefore less prone to TDD than are transcripts with median-to-high ribosome densities. Importantly, similar results are obtained whether cycloheximide (which blocks ribosomes on the coding sequence) or harringtonine (which allows 80S ribosome run-off from the coding sequence) is used to inhibit translation (Fig. 2B; Supplemental Fig. S5B). This indicates that the observed relationship between ribosome density and TDD does not result from the physical protection of the coding region by ribosomes stalled by the cycloheximide treatment. Moreover, similar results were obtained when using the fold-change in transcript abundance upon blocking translation with cycloheximide or harringtonine alone (in the absence of transcription inhibitors) as a proxy for the TDDindex, thus confirming that our observations do not result from a technical bias introduced by the use of transcription inhibitors (Supplemental Fig. S6A–D).
Exon count also appears as an important transcript feature that is positively associated with TDD (Fig. 2A,B). This link does not appear to be driven by transcript length (Fig. 2B), therefore suggesting a specific role of exon–exon junctions in regulating TDD. Contrary to exon count, the occurrence of the AU-rich core pentamer motif (AUUUA) in the 3′ UTR of cellular transcripts is negatively correlated with the TDDindex (Fig. 2A,B). AU-rich elements are able to recruit specific RNA-binding proteins involved in modulating mRNA stability (Chyi-Ying and Ann-Bin 1995). When looking at the extended AU-rich motif (WWWAAUUUAAWWW), which is the functional unit required to induce mRNA decay, we observe that transcripts bearing this extended motif have a significantly smaller TDDindex (Wilcoxon P < 2.2 × 10−16 and Cohen's d = 0.53) than the overall transcript population (Supplemental Fig. S5C). Our results therefore indicate that AU-rich mediated mRNA decay does not occur through a translation-dependent mechanism.
Our results also indicate a global anticorrelation between the secondary structure in the coding region and the TDDindex (Fig. 2B). Transcripts with highly structured CDS are more prone to TDD than are transcripts with less-structured CDS (Fig. 2B). This is in agreement with a previous report indicating a role for secondary structure in the coding region in modulating TDD (Mauger et al. 2019).
Finally, m6A modifications have been linked to the regulation of mRNA translation and stability (Meyer 2019; Lee et al. 2020), although this feature did not appear as a major factor connected to the extent of TDD. However, this could be because of a bias introduced by the fact that a large fraction of mRNAs do not contain detectable m6A sites. We therefore decided to compare the distribution of TDDindexes among transcripts containing or not detectable m6A sites in different transcript regions (i.e., 5′ UTR, around the start and stop codons, in the CDS and 3′ UTR). As shown (Supplemental Fig. S5D), the presence of m6A sites in the 5′ UTR is associated with a mild but significant decrease in TDD (Wilcoxon P = 4.4 × 10−6, Cohen's d = 0.19). m6A sites in other regions do not appear to modulate the extent of TDD or suffer from small effect size as is the case for m6A sites in the CDS (Wilcoxon P = 1.1 × 10−5, Cohen's d = 0.08).
Taken together, our results suggest that TDD is a complex process that is modulated by multiple cis- and trans-acting features. Among these, the length of UTRs and the density of ribosomes across the coding sequence appear as major factors in mediating or modulating the extent of TDD, together with other factors such as the extent of secondary structure in the coding sequence and the number of exons.
mRNA features defining the extent of TID in T CD4+ lymphocytes
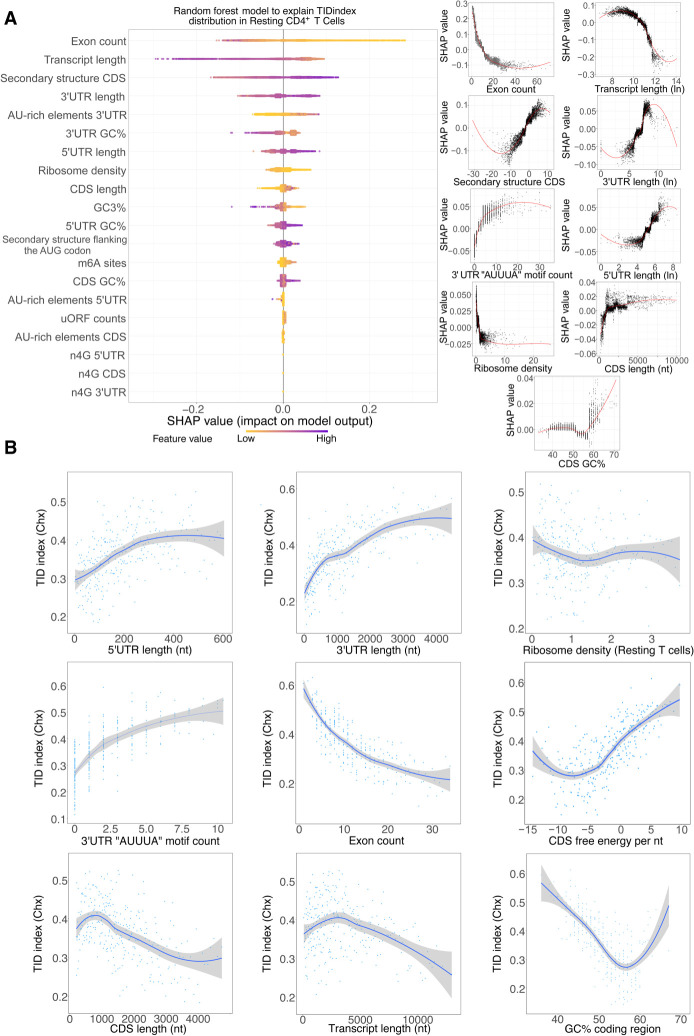
When applied to the TIDindex, random forest analysis (Supplemental Code S1) was able to explain 40% of the observed variance (for plots of predicted vs. observed TIDindex, see Supplemental Fig. S7A). In this case, the number of exons per transcript and the total transcript length were the two most important factors able to predict the extent of TID in resting T cells (Fig. 3A). Again, similar results were obtained when using harringtonine to inhibit translation (Supplemental Fig. S7B). The TIDindex shows a strong anticorrelation with the number of exons per transcript (Fig. 3B). Consistent with this observation, long transcripts (which usually have a large number of exons) tend to be less dependent on TID for their decay (Fig. 3B). However, among shorter transcripts, there is no clear trend and rather a small positive correlation between transcript length and the extent of TID (Fig. 3B). This, together with the fact that the length of the CDS could be positively correlated to the TIDindex, as shown in the SHAP value plot (Fig. 3A, right), suggests that the effect of exon number on TID is independent from the total length, consistent with previously published data indicating a correlation between exon junction density and mRNA stability that could be driven by masking of m6A sites by the exon–junction complex upon splicing (Spies et al. 2013; Agarwal and Kelley 2022; He et al. 2023; Uzonyi et al. 2023).
Figure 3.
Characterization of cis- and trans-acting features linked to TID. (A) Random forest decision tree analysis of transcript features to explain the observed TIDindex values in resting CD4+ T lymphocytes coupled with SHAP value analysis for interpreting the output of the random forest model. (Left) Features are sorted from top to bottom with respect to their importance in predicting the TDDindex and SHAP values displayed to show how much each feature has influenced the model. (Right) Plots showing the impact of SHAP values on the model for the main identified features. (B) Binning plots of the TIDindex against selected features. Transcripts are first ordered with respect to the feature to be compared with the TIDindex and groups of 20 transcripts made along the selected feature. The mean TIDindex and feature values are plotted for each group.
The length of UTRs appears to be linked to the extent of TID. Indeed, transcripts bearing long 3′ UTRs and 5′ UTRs are more prone to TID than those with short UTRs (Fig. 3B). Furthermore, there is a relationship between the degree of secondary structures in the CDS and the extent of TID. Transcripts with highly structured coding sequences tend to be less degraded in a translation-independent manner than those with low CDS secondary structure (Fig. 3A,B).
AU-rich elements in the 3′ UTR are positively correlated with the TIDindex (Fig. 3B; Supplemental Fig. S7C), further corroborating that this active mRNA decay pathway, mediated by specific RNA-binding proteins, mainly occurs in a translation-independent manner.
Finally, although ribosome density is ranked among the important predictors of TID by the random forest model, we do not observe a clear relationship between ribosome density and the extent of TID in the binning plot (Fig. 3B). However, the SHAP value plot corresponding to ribosome density does show a clear negative correlation with the TDDindex (Fig. 3A). This suggests that the effect of ribosome density on the observed TID among the transcriptome could be masked in the binning plot by other important features responsible for modulating TID (such as exon count, UTR length, etc.).
Taken together, our results indicates that TID strongly regulates transcripts with few exons, long UTRs, and AU-rich elements in their 3′ UTR and with possibly low ribosome occupancy.
Activation of primary mouse CD4+ T cells induces profound transcriptome and translatome remodeling with concomitant changes in mRNA stability
Having characterized the features favoring TDD and TID in resting T cells, we tested whether T cell activation could induce changes in these relationships. Indeed, upon activation, T cells undergo an extensive change in their gene expression program and metabolism to exit quiescence and enter into a proliferative state.
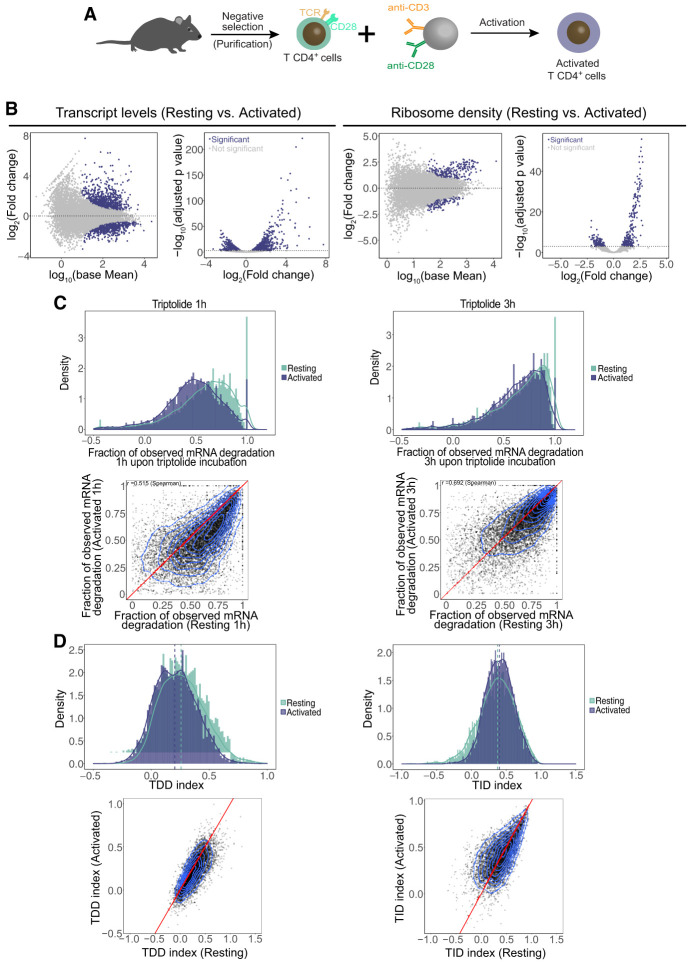
Primary mouse CD4+ T cells were activated with anti-CD3/anti-CD28 antibody-coated beads, mimicking antigen-presenting cells (Fig. 4A). Activation was confirmed by strong cell surface expression of activation markers, as well as cellular proliferation (Supplemental Fig. S8A). As with resting T cells, we prepared whole-cell RNA-seq and ribosome footprinting libraries from three independent biological replicates of T cells after 3 h of activation (activated). As expected, anti-CD3/CD28 stimulation significantly altered the T cell transcriptome (Fig. 4B; Supplemental Table S1). Overall, 822 transcripts showed a significant increase in expression upon activation, and 694 transcripts exhibited decreased expression (Fig. 4B, left). GO analysis indicates that differentially up-regulated transcripts at 3 h after activation include pathways related to gene expression regulation such as ribosome biogenesis, translation initiation, tRNA processing, mRNA processing, and regulation of RNA polII transcription, as well as pathways related to immune system and response to cytokine (Supplemental Fig. S8B; Supplemental Table S1). Significantly down-regulated transcripts are enriched for ribosomal proteins, protein ubiquitination factors, and factors involved in the cell cycle, T cell differentiation, and RNA splicing among others (Supplemental Fig. S8C; Supplemental Table S1).
Figure 4.
Changes in RNA abundance, ribosome density, and mRNA decay rates induced by T cell activation. (A) Schematic representation of the procedure for activation of primary CD4+ T lymphocytes. (B) Differential gene expression analysis of transcript abundance as measured by RNA-seq (left) and ribosome density as measured by ribosome profiling (right) in resting and activated cells. (C) Fraction of mRNA degradation in resting and activated T cells 1 and 3 h after transcription inhibition with triptolide. (D) Comparison of the distribution of the TDDindex (left) and TIDindex (right) in resting and activated T cells.
Similarly, anti-CD3/CD28 stimulation altered the translatome of T cells (Fig. 4B, right). Ribosome profiling revealed hundreds of transcripts displaying significant changes in ribosome density in their coding sequence upon T cell activation (Fig. 4B, right). Those displaying a significant increase in ribosome density are enriched for ribosomal proteins (78 out of the 80 core ribosomal proteins), ribosome biogenesis factors, RNA splicing factors, and factors involved in the cellular response to interleukin 4 (Supplemental Fig. S8D; Supplemental Table S1). Conversely, transcripts displaying a significant decrease in ribosome density mainly code for kinases, signal transduction factors, and transcription factors (Supplemental Fig. S8E; Supplemental Table S1).
Comparison of the fraction of observed mRNA degradation () at 1 h and 3 h upon triptolide addition in both resting and activated T cells (Fig. 4C) indicates that mRNA decay is globally more efficient in resting cells compared with activated cells. This result is consistent with a recent report describing a global stabilization of mRNAs upon CD4+ T cell activation in mice (Hwang et al. 2020). When breaking apart mRNA degradation into TDD and TID, it appears that T cell activation triggers a decrease in TDD levels, whereas overall TID is slightly increased (Fig. 4D).
T cell activation inverts the relationship between GC3 content and TDD/TID
Random forest analysis performed with the TDDindex and TIDindex in activated cells yielded similar results to those in resting cells, except that AU-rich elements in the 3′ UTR became a less important predictor of both TDD and TID in activated cells (Fig. 5A,B). Conversely, the percentage of GC in the coding region and GC3 content (GC content at the third position of codons) became a better predictor of both TDD and TID in activated cells (Fig. 5A,B).
Figure 5.
Comparison of features associated with TDD in resting and activated CD4+ T lymphocytes. (A,B) Random forest decision tree analysis (with SHAP value analysis for interpreting the output of the random forest model) of transcript features to explain the observed TDDindex (left) and TIDindex (right) values in activated CD4+ T lymphocytes. Features are sorted from top to bottom with respect to their importance in predicting the TDDindex or TIDindex. (C,D) Binning plots of TDDindex (left) or TIDindex (right) against selected features. Transcripts are first ordered with respect to the feature to be compared with TIDindex and groups of 20 transcripts made along the selected feature. The mean TIDindex and feature values are plotted for each group.
Binning plots of 3′ UTR length, 5′ UTR length, and ribosome density against the TDDindex revealed similar relationships in activated T cells than those observed in resting cells (Fig. 5C,D). The relationship between secondary structure in the coding sequence and the TDDindex was lost upon T cell activation (Fig. 5C,D). Moreover, GC content in the coding region, which is positively correlated to the TDDindex in resting T cells (Fig. 2B), displays the opposite trend upon T cell activation (see Fig. 5C,D).
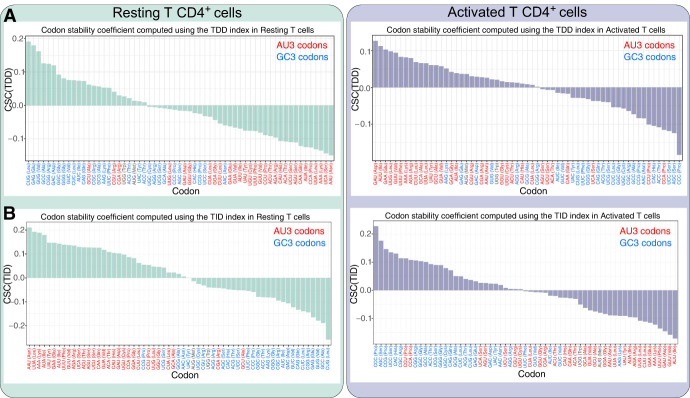
Because our data sets allow discrimination of mRNA degradation into TDD and TID, we tested the relationship between codon usage and the two mRNA decay pathways. Codon usage and transcript stability have been recently studied using a specific metric known as the codon occurrence to mRNA stability correlation coefficient (CSC) (Presnyak et al. 2015). The CSC is defined as the Pearson's correlation between the frequency of each codon in mRNAs and the half-lives of the mRNAs. Here, rather than calculating a correlation with mRNA half-lives, we implemented the TDD-CSC and TID-CSC metrics by calculating the Pearson's correlation between codon frequency and the TDD or TID indexes, both in resting and activated T cells. TDD-CSC and TID-CSC values are within similar ranges (between −0.2 and 0.2) (see Fig. 6A,B) to those obtained from the literature using mRNA half-lives as input (Presnyak et al. 2015; Wu et al. 2019; Forrest et al. 2020; Gillen et al. 2021). Consistent with the random forest analysis, in resting T cells, we could clearly see a nucleotide bias within codons bearing positive or negative TDD-CSC and TID-CSCs (Fig. 6A,B, left panels). Regarding TDD, GC-rich codons are mostly associated with positive TDD-CSCs in resting cells (i.e., GC-rich codons are more frequent within transcripts highly degraded by TDD), whereas AU-rich codons are mainly associated with negative TDD-CSCs (i.e., AU-rich codons are more frequent within transcripts poorly degraded by TDD) (Fig. 6A, left). GC-rich codons are most frequently associated with negative TID-CSCs (i.e., GC-rich codons are more frequent within transcripts poorly degraded by TID), whereas AU-rich codons typically display positive TID-CSCs (i.e., AU-rich codons are more frequent within transcripts highly degraded by TID) (Fig. 6B, left). This observation was even more pronounced when looking at the GC content of the third position of codons (GC3) (Fig. 6A,B, left; Supplemental Fig. S9A,C). Notably, for TDD, 21 out of 24 codons with a positive TDD-CSC value ended with a G or C (Fig. 6A,B, see blue codons; Supplemental Fig. S9A, left), whereas only 10 out of 37 codons with a negative TDD-CSC value ended with a G or C (Supplemental Fig. S9A). This distribution is significantly biased from a random distribution among codons with positive and negative CSC(TDD) values (P-value = 1.34 × 10−5, chi-squared test) (Supplemental Fig. S9A, left). For TID, only six out of 33 codons with a positive TID-CSC ended with a G or C, whereas 25 out of 28 codons with a negative TID-CSC ended with a G or C (P-value = 1.30 × 10−7, chi-squared test) (Supplemental Fig. S9C, left). Upon T cell activation, the relationship between codon GC content, TDD, and TID is flipped, leading to the GC3-rich codon being mainly associated with negative TDD-CSCs (P-value = 0.0012, chi-squared test) (Supplemental Fig. S9A) and positive TID-CSCs (P-value = 0.0002, chi-squared test) (Supplemental Fig. S9C, right), whereas AU-rich codons (Fig. 6A,B, see red codons) are mainly associated with transcripts degraded by TDD, bearing positive TDD-CSCs and negative TID-CSCs (Fig. 6A,B; Supplemental Fig. S9B,D).
Figure 6.
Relationship between GC content in the coding sequence, TDD, and TID. (A) Codon stability coefficient computed for each codon using the TDDindex (CSC(TDD)) in resting (left, green bars) and activated (right, violet bars) T CD4+ cells. Codons colored in red correspond to AU3 codons, and codons colored in blue correspond to GC3. (B) Same as A but using the TIDindex (CSC(TID)) instead of the TDDindex.
Our results therefore suggest that GC content in the coding sequence and particularly at the third position of codons (GC3) is an important determinant for translation dependence. Moreover, the differences observed between resting and activated T cells further suggest a dynamic relationship between GC content in the coding sequence and mRNA decay.
TDD and TID are competing pathways defined by changes in ribosome density during T cell activation
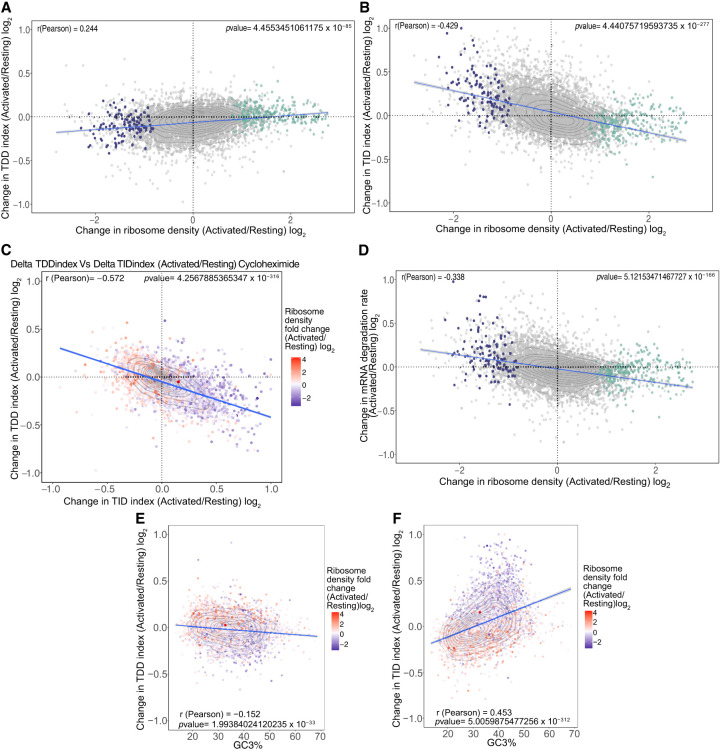
Our results suggest that ribosomes themselves could act as triggers of TDD (Fig. 2B), the extent of which is modulated by transcript features such as 3′ UTR length, GC content in the coding region, or exon number. As a consequence, any change in ribosome density should lead to a corresponding change in the observed TDDindex. To test this, we plotted for each transcript the change in TDD index between activated and resting T cells against the corresponding fold-change in ribosome density (Fig. 7A). We observed a moderate but significant positive correlation (r = 0.244) between changes in ribosome density and changes in the TDDindex for the entire transcriptome. The same observation was made whether cycloheximide or harringtonine was used to block translation in resting and activated T cells (cf. Fig. 7A and Supplemental Fig. S10A). Transcripts displaying an increase in their ribosome density upon T cell activation tend to display a concomitant increase in their TDDindex, whereas transcripts displaying a decrease in their ribosome density tend to display a decrease in their TDDindex. To test whether changes in TDDindex could also be driven by other transcript features, we applied random forest analysis to rank the features that could explain the extent of changes in both indexes upon T cell activation (for model prediction and feature importance, see Supplemental Fig. S10D,E). As expected, changes in ribosome density ranked among the best variables explaining changes in the TDDindex, together with GC content in the coding sequence (Supplemental Fig. S10D). The fact that the GC content in the coding sequence impacts differently the TDDindex upon T cell activation agrees with our previous observation of an inversion in the relationship between TDDindex and GC3 content upon T cell activation (see Fig. 6). Indeed, transcripts with high GC3 content in their CDS display a decrease in their TDDindex, and transcripts with low GC3 content in their CDS display an increase in their TDDindex upon T cell activation (Fig. 7E).
Figure 7.
Changes in ribosome density upon T cell activation modulate both TDD and TID. (A) Scatter plot of the changes in ribosome density between resting and activated cells (x-axis) against the changes in the TDDindex (y-axis). Each dot corresponds to a single transcript. Violet and green dots correspond to transcripts from Figure 1D displaying a significant decrease (violet) or increase (green) in ribosome density upon T cell activation. (B) Scatter plot of the changes in ribosome density between resting and activated cells (x-axis) against the changes in the TIDindex (y-axis). (C) Scatter plot of the change in TDDindex (y-axis) and TIDindex (x-axis) between resting and activated cells. Transcripts are colored with respect to the change in ribosome density measured between resting and activated cells. (D) Scatter plot of the changes in ribosome density between resting and activated cells (x-axis) against the changes in mRNA degradation rate (y-axis). (E,F) Scatter plot of the GC3% against changes in TDDindex and TIDindex between resting and activated cells.
Having identified changes in ribosome density as well as GC3 content in the CDS as important factors driving observed changes in the TDDindex upon T cell activation, we performed a similar analysis to identify factors driving changes in the observed TIDindex (Supplemental Fig. S10E, right). Changes in ribosome density were also among the main factors explaining the observed changes in the TIDindex upon T cell activation, together with the CDS GC and GC3 content (Fig. 7F; Supplemental Fig. S10E, right). In this case, changes in ribosome density display a negative correlation against changes in TIDindex (r = −0.429) (see Fig. 7B). When compared together, changes in TDD and TID upon T cell activation are anticorrelated and appear to be linked to changes to ribosome density (Fig. 7C). Finally, as previously observed, the overall contribution of TID in total mRNA decay is preponderant over that of TDD. As a consequence, total mRNA degradation is negatively correlated to changes in ribosome density upon T cell activation (Fig. 7D). mRNAs with increased ribosome density upon T cell activation are stabilized, whereas those that display a reduction in ribosome density upon T cell activation are destabilized (Fig. 7D).
Taken together, our results suggest that TDD and TID occur through mutually exclusive pathways that compete for the same substrate and are defined by ribosome density as well as transcript cis-acting features that participate in modulating the efficiency of each pathway.
Discussion
TDD has emerged as an important determinant of mRNA stability among eukaryotes. However, its precise contribution to overall mRNA decay and the features that modulate its efficiency are still poorly characterized. Using a combination of transcription and translation inhibitors, we were able to deconvolute mRNA decay into translation dependent and translation independent (Fig. 1). Our results uncover a global effect of mRNA translation in inducing mRNA decay (Fig. 1B). Furthermore, the extent of TDD appears to be a transcript-specific feature, with most cellular transcripts displaying moderate TDD (median of 27% of observed mRNA decay at 3 h being explained by a translation-dependent mechanism), whereas others are either entirely degraded through TDD or completely insensitive to it (Fig. 1F). As expected, known TDD targets such as UPF2-regulated transcripts are highly dependent on translation for their decay, whereas lincRNAs are not (Fig. 1E). Random forest analysis of transcript cis- and trans-acting features allowed the identification of different features involved in mediating or modulating TDD and TID in resting and activated T cells (for a global overview, see Supplemental Fig. S11). However, it is important to notice that the list presented here is not exhaustive and that other potential regulators of TDD and TID such as RNA-binding proteins and microRNAs were not taken into account in this study.
Our results revealed that ribosome density is an important determinant of TDD both in resting and activated T cells (Figs. 2, 5, left panels). The relationship between TDD and ribosome density is monotonic followed by saturation, with a positive linear correlation between TDD and ribosome density for low-to-medium ribosome density values, which reaches a plateau for higher ribosome density values (Fig. 2B). These results are in agreement with a progressive mRNA decay model recently proposed by Dave et al. (2023), using single-molecule analysis of TDD. This model suggests that ribosome flux, which depends on the translation-initiation frequency, is responsible for mRNA destabilization rather than the total number of ribosomes on a given mRNA. Moreover, the model predicts a nonlinear relationship between mRNA degradation and translation-initiation rates, showing a plateau at fast initiation rates similar to what we observe with the TDDindex for mRNAs with high ribosome density (Fig. 2B). Altogether, it is tempting to speculate that ribosomes themselves could be the main factor responsible for inducing TDD, most likely via recruitment of specific decay factors. This hypothesis is supported by evidence showing that ribosomes are hubs for the assembly of factors involved in mRNA surveillance and have been shown to directly interact with different mRNA decay factors (Simsek et al. 2017; Tesina et al. 2019; Buschauer et al. 2020; Lin et al. 2020; Tuck et al. 2020). Furthermore, the frequency of “benign” ribosome collisions has been recently reported to be positively correlated to ribosome density in a transcriptome-wide manner (Arpat et al. 2020) and, in some instances, to recruiting of the RNA decay factor SKIV2L (Tuck et al. 2020). This suggests that ribosome collisions could be responsible for the correlation between ribosome density and TDD that we observe in our data sets.
Our random forest analysis also pointed to transcript 3′ UTR length and, to a lesser extent, to 5′ UTR length among important factors explaining TDD (Fig. 2A). Further analysis confirmed a global anticorrelation between UTR length and TDD susceptibility (Fig. 2B). This result was surprising because long 3′ UTRs had been linked to UPF1-dependent mRNA decay (Hogg and Goff 2010; Kurosaki and Maquat 2013). However, there is evidence that the role of UPF1 in the decay of long 3′ UTRs could occur through a ribosome-independent pathway involving the microRNA pathway and the CCR4-NOT deadenylase complex (Park et al. 2019), as well as independently from NMD within chromatoid bodies through TDRD6 (Fanourgakis et al. 2016). More recently, nanopore sequencing of cellular transcripts from cells in which NMD has been inhibited did not find any correlation between 3′ UTR length and NMD susceptibility (Karousis et al. 2021). Furthermore, in zebrafish, 3′ UTR length has been shown to play a buffer role against deadenylation and decay of maternal mRNAs mediated by codon usage (Mishima and Tomari 2016). In this pathway, long 3′ UTRs were shown to decrease accessibility of mRNA decay factors recruited by nonoptimal codons to the 3′ poly(A) tail (distance model of codon-dependent mRNA decay). These recent studies are consistent with our findings.
Studies performed in budding yeast (Saccharomyces cerevisiae) have pointed to codon usage coupled to ribosome residency time as features conditioning recruitment of mRNA decay factors to ribosomes, thus leading to TDD (Presnyak et al. 2015; Radhakrishnan et al. 2016). Similar results have been found in higher eukaryotes, in which enrichment in specific codons was shown to lead to higher mRNA decay rates (Narula et al. 2019; Wu et al. 2019; Forrest et al. 2020). GC and GC3 content along the coding sequence have also been described to drive TDD in human cells (Hia et al. 2019) in a context in which GC3-rich codons tend to destabilize mRNAs whereas AU3-rich codons stabilize them. Such a GC3 bias can also be observed in the codons associated with stable and unstable mRNAs from previous studies (Narula et al. 2019; Wu et al. 2019; Shu et al. 2020), although this observation was not directly highlighted by the investigators. In resting T cells, our results show that TDD is mainly associated with GC3-rich codons, whereas TID is associated with AU3-rich codons (Fig. 6A,B). Because TID accounts for a larger fraction of mRNA decay than does TDD, its effects predominate when assessing mRNA degradation rates and codon usage. Therefore, it introduces a bias when mRNA stability is used as a proxy to study TDD pathways. Finally, as we observed a global inversion of the observed relationship between GC content in the CDS, TDD, and TID upon T cell activation (Fig. 6), our results further suggest that this relationship can be dynamically regulated, possibly through the modulation of trans-acting factors. Loss of FMRP in mouse neurons leads to reshuffling of the link between codon usage and mRNA stability (Shu et al. 2020) similar to the one we observe upon T cell activation. It suggests that T cell activation might be accompanied by changes in the relative levels or activity of specific RNA-binding proteins involved in the recognition of specific codons or GC content in the CDS. Consistently, it has been recently shown that codon usage, tRNA abundance, and editing are dynamically regulated in mouse CD4+ T cells (Rak et al. 2021; Liu et al. 2022). In this particular context, expression of mRNAs enriched in “proliferation codons” bearing a strong AU3 bias tends to be up-regulated 20 h upon CD4+ T cell activation, whereas expression of mRNAs enriched in “differentiation codons” bearing a strong GC3 bias tends to be down-regulated. These changes are accompanied by corresponding changes in tRNA abundance that could compensate for the imbalance in codon usage (Rak et al. 2021). This observation could explain the switch that we observe with TDD and TID with respect to the GC3 bias in codon usage, as well as the increase in ribosome density for transcripts bearing AU3-rich coding regions (Fig. 7F).
Finally, in resting and activated T cells, TDD mainly targets transcripts coding for proteins with basic functions related to the process of gene expression, which are generally abundant in the cell. These include ribosome biogenesis factors, factors implicated in tRNA metabolism, mRNA splicing, and the proteasome. For those transcripts, TDD could participate in limiting the protein output per mRNA unit and introduce a negative feedback loop to avoid protein overexpression. Such a feedback loop would be particularly useful under conditions in which expression is up-regulated, as observed for several of these GO categories upon T cell activation (Supplemental Fig. S8), when cell volume increases rapidly, which requires an increase in protein levels to prepare for clonal expansion. TDD could thus represent an additional level of regulation for types of proteins in which overexpression could be detrimental for T cells. This could be useful for key component of the TCR signaling (such as transcripts encoding CD3 chains and ZAP that specifically rely on TDD for their decay), because any dysregulation in those pathways can lead to abnormal immune responses.
Conversely, TID predominantly targets functional categories of transcripts that are related to the fine-tuning of gene expression (transcription factors) or specific cellular pathways such as cell cycle and the immune response. These pathways require a dynamic and reversible control of gene expression and mostly rely on specific trans-acting factors (such as AU-rich binding proteins) to actively regulate their expression both temporally and in their amplitude. Transcripts highly regulated by TID display long 3′ UTRs (Fig. 3B), therefore providing a larger platform to recruit RBPs involved in the post-transcriptional regulation of gene expression (Sandberg et al. 2008; Gruber et al. 2014).
Methods
Primary cell purification and culture
Primary CD4+ T cells were obtained from 6-wk-old C57BL/6J female mice. Briefly, the spleen as well as the inguinal, axillary, brachial, cervical, and mesenteric lymph nodes were collected, followed by Ficoll separation to remove red blood cells from splenocytes. CD4+ T cells were then purified by negative selection using the CD4+ T cell isolation kit (Miltenyi Biotec 130-104-454) following the manufacturer's protocol. Isolated cells were grown in RPMI medium supplemented with 10% fetal calf serum (FCS) and 50 µM β-mercaptoethanol. CD4+ T cell activation was performed using magnetic beads coupled with CD3/CD28 antibodies (Thermo Fisher Scientific 11452D) following the manufacturer's protocol.
Cell viability, proliferation, and cell surface marker detection by flow cytometry analysis
Before culture, CD4+ T cells were stained with carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes). After 24, 48, or 72 h of culture, cell numbers were measured by flow cytometry using Calibrite beads (BD Pharmingen) as standards as previously described (Cottalorda et al. 2006). Expression of cell surface markers in resting and CD3/CD28-activated cells was made by flow cytometry using fluorescent-coupled antibodies against CD4 (Biolegend 100434), CD3 (Biolegend 100308), CD69 (Biolegend 104507), and CD25 (Biolegend 102021).
RNA stability measurements
To monitor mRNA stability, 3 million CD4+ T cells were incubated in the presence of DRB (Sigma-Aldrich D1916) at a final concentration of 65 µM or triptolide (Sigma-Aldrich T3652) at a final concentration of 25 µM for 15 min, 1 h, or 3 h. At each time point (0, 15 min, 1 h, and 3 h), cells were collected and counted, and total RNA was extracted from the same number of cells (3 million) at each time point using TRIzol in the presence of 1 µL of a 1/10 dilution of ERCC spike-in RNA (Thermo Fisher Scientific 4456740). To monitor mRNA stability in conditions in which mRNA translation is impaired, cells were incubated in the presence of either DRB or triptolide and cycloheximide (final concentration of 100 µg/mL; Sigma-Aldrich 01810) or harringtonine (final concentration of 2 µg/mL; Interchim H0169) for 1 or 3 h and total RNA extracted as described above. Total RNA was depleted from ribosomal RNA using the Ribo-Zero rRNA removal kit (human/mouse/rat, Illumina) followed by cDNA library preparation as described below. All experiments were performed in three independent biological replicates obtained from mouse primary CD4+ T cells purified from independent groups of 6-wk-old C57BL/6J female mice on different days.
For 4-thiouridine labeling experiments, cells were incubated for 24 h in the presence of the indicated concentration of 4-thiouridine in RPMI medium supplemented with 10% FCS and 50 µM β-mercaptoethanol. The medium was changed every 3 h to maintain 4-thiouridine levels at the indicated concentration. After 24 h, cells were labeled with eBioscience Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific 65-0865-18) according to the manufacturer's recommendations, fixed with PSB-PFA 4%, and analyzed by flow cytometry on a BD LSRII instrument.
RNA-seq cDNA library preparation
High-throughput sequencing libraries were prepared as previously described (Heyer et al. 2015). More information can be found in the Supplemental Methods.
Ribosome profiling
Ribosome profiling libraries were prepared as previously described (Ricci et al. 2014). More information can be found in the Supplemental Methods.
Poly(A)-site sequencing
Poly(A)-site sequencing libraries were prepared as previously described (Ashar-Patel et al. 2017). More information can be found in the Supplemental Methods.
Analysis of high-throughput sequencing reads
All the scripts used for this analysis are available at the following repository: https://gitbio.ens-lyon.fr/LBMC/RMI2/tdd_project.
Sequencing reads were split with respect to their 5′ in-line barcode sequence. After this, 5′-barcode and 3′-adaptor sequences were removed from reads using FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Reads were mapped to a custom set of sequences corresponding to the mouse 18S, 28S, 45S, 5S, and 5.8S rRNA; tRNAs; and the ERCC spike-in sequences (available in Supplemental Table S2) to filter them out, using Bowtie 2 (version 2.2.4) (Langmead and Salzberg 2012) with the following parameters “Bowtie 2 -t ‐‐fast”.
For RNA sequencing and ribosome profiling samples, reads that failed to map to this custom set of sequences were next aligned to the mouse mm10 assembly and the GENCODE vM7 annotation using TopHat2 (v2.0.13) (Kim et al. 2013) with the following parameters: “Tophat2 ‐‐Bowtie (Bowtie version 1.1.1.0) ‐‐library-type fr-secondstrand ‐‐b2-sensitive -i 30 -m 1 -g 10 ‐‐max-coverage-intron 1000000”. Read counts on all transcripts of interest were obtained using the HTSeq count package (Anders et al. 2015) with the following parameters “htseq-count -f sam -r pos -s yes -a 10 -m intersection-nonempty”.
Transcript database creation
The script to generate the transcript database and its documentation can be found in Supplemental Code S2 and https://gitbio.ens-lyon.fr/LBMC/RMI2/rmi2_gff_fasta_compilation. The database can be found in Supplemental Table S3. The rationale to build the database was to eventually select a single transcript isoform to quantify expression and ribosome density for further analyses and obtain transcript information such as the length of UTRs, coding sequence, codon usage, and other features used to build the random forest model. Additional information regarding how the transcript database was built can be found in the Supplemental Methods section.
Differential gene expression analysis with DESeq2
Differential expressed genes and translated genes upon cell activation were obtained using DESeq2 (version 1.24.0) (Love et al. 2014) in R (version 4.3.2) (R Core Team 2023) using, respectively, RNA-seq and ribosome profiling read counts of activated T cells versus resting T cells with a classical model design ∼ condition + replicate.
To obtain genes that differ in ribosome density, a DESeq2 model was constructed using both RNA-seq and ribosome profiling read counts with the following model:
∼condition:sequencing + condition + replicate:sequencing,
where condition refers to the resting/activated state of the cells and sequencing to RNA-seq/RiboSeq libraries. Specifically changes in ribosome density were recovered using the contrast
list (“conditionactivated.sequencingRiboSeq”, “conditionresting.sequencingRiboSeq”) of the results command.
Read count normalization by DESeq with endogenous stable genes
To define endogenous stable genes, read counts normalized using the exogenous spike-in reads were plotted to visually identify stable transcripts. Based on this information, the 0-h triptolide versus 3-h triptolide in read per million (RPM) was plotted to check that the selected transcripts were indeed relatively enriched in the 3-h triptolide condition compared with the control condition for all biological replicates.
Stable genes were then used by DESeq2 (1.24.0) (Love et al. 2014) to estimate the size factor of all libraries, and then normalized read counts were recovered in a matrix for downstream analysis and calculation.
Functional enrichment analysis of transcripts differentially expressed and translated upon cell activation
Functional enrichment analysis was performed with DAVID (version 6.8) (Da Huang et al. 2009a,b; https://david.ncifcrf.gov/summary.jsp), selecting genes with an adjusted P-value < 0.05 in the differential gene expression and differential translation analysis. Plots were generated with Revigo (Supek et al. 2011; http://revigo.irb.hr/) using as inputs GO terms with an adjusted P-value < 0.01 as calculated by the DAVID software.
Random forest models
Scripts to perform random forest models can be found in Supplemental Code S1. First, the gene set of interest was reduced to the expressed genes based on the gene normalized read counts of 3-h triptolide libraries (about 5000 genes in resting and about 6000 genes in activated T cells). Of these, only genes with completed observations in all biological replicates (including ribosome profiling libraries) and for all transcript features used to build the model were kept. The data set was then divided in a train and validation data set composed of, respectively, 80% and 20% genes randomly chosen. For model development, XGBoost, was used in R (version R 4.3.2). The train function from the caret (v6.0-94) package was used. The train function iteratively refits the model over bootstrap samples and explores various options for the number of randomly selected predictors at each split in the tree (controlled by the colsample_bytree parameter). To evaluate the model's performance, predictions were made on the validation data set. The Spearman's correlation coefficient was then calculated to assess the concordance between the predicted and observed values. Following model training, SHAP values were computed in order to identify feature importance using the SHAPforxgboost (v 0.1.3; https://github.com/liuyanguu/SHAPforxgboost) package.
The accuracy of the final model obtained by the train function was then verified by predicting the parameters of interest (TDDindex or TIDindex) of the validation data set with the random forest model and calculating the Spearman's correlation between the predicted versus the real value.
G-quadruplex prediction
G-quadruplex structures within transcripts were predicted as previously described (Beaudoin et al. 2014) using a dedicated Microsoft Excel macro available in Supplemental Code S3.
RNA secondary structure modeling
RNA secondary structure modeling in the 5′ UTR, CDS, and 3′ UTR was performed using the ViennaRNA package (version 2.5.1) (Lorenz et al. 2011). A normalized minimum free energy score (MFE) for each sequence was then calculated (i.e., MFEden) as previously described (Trotta 2014).
CSC correlation–permutation sequence
CSC scores were calculated as inspired by Presnyak et al. (2015). Briefly, the CSC score is the Pearson's correlation between the frequency of occurrence of each codon and the TDDindex of the corresponding transcript. Statistical significance was determined by permutating 10,000 times the sequence of each coding sequence. This allow us to generate 10,000 transcriptomes with random codons from transcripts that share same features as the original sequence (GC content, CDS length). For each random transcriptome, the CSC score for each codon was computed, allowing us to calculate a CSC score distribution obtained randomly. The P-value corresponds to the probability of obtaining the observed CSC score from the randomized transcriptome. False-discovery rates (FDRs) were calculated with the Benjamini and Yekutieli method (Benjamini and Yekutieli 2001) to adjust the P-value for multiple comparison tests.
GO analysis using TDDindex and TIDindex values
Transcripts were associated to GO terms using the mgi.gaf file (http://current.geneontology.org/annotations/mgi.gaf.gz; downloaded September 17, 2019); the GO terms to GO phrase association, as well as the GO tree, were obtained from the go-basic.obo file (http://purl.obolibrary.org/obo/go/go-basic.obo; downloaded September 17, 2019). The distributions of TDDindex and TIDindex values were generated for each GO and compared with the distribution obtained for the global transcript population. The statistical significance of the difference in the mean TDDindex or TIDindex value between the transcripts from any given GO and the global population was determined by a bootstrapping test (50,000 random ensembles of same dimension of the GOs). The obtained P-values were then submitted to a hierarchical FDR-controlling methodology (Benjamini and Yekutieli 2001), using the structure of the GO tree.
Quantitative PCR analysis of mRNA half-lives
To monitor mRNA half-lives, 3 million CD4+ T cells were incubated in the presence of triptolide (Sigma-Aldrich T3652) at a final concentration of 25 µM for 1 h, 2 h, 3 h, 4 h, 5 h, or 6 h. At each time point, cells were collected and counted, and total RNA was extracted from the same number of cells (3 million) at each time point using TRIzol in the presence of 0.5 ng of an in vitro transcribed cap and polyadenylated Renilla luciferase mRNA. To monitor mRNA stability in conditions in which mRNA translation is impaired, cells were incubated in the presence of triptolide and cycloheximide (final concentration of 100 µg/mL; Sigma-Aldrich 01810) and total RNA extracted at each time point as described above.
cDNA was generated using the high-capacity cDNA reverse transcription kit (Applied Biosystems) with 200 ng of total RNA, according to the manufacturer's instructions.
qPCRs were performed with SensiFAST SYBR no-rox kit (Bioline) using the CFX connect real-time PCR detection system.
Thermal cycling parameters included a 2-min preincubation at 95°C and 40 amplification cycles for 10 sec at 95°C and for 30 sec at 60°C. All sequences of primers are described in Supplemental Table S4.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE159301. All the scripts used for the analyses presented in the paper are available as Supplemental Code and at the following repositories: https://gitbio.ens-lyon.fr/LBMC/RMI2/tdd_project and https://gitbio.ens-lyon.fr/LBMC/RMI2/rmi2_gff_fasta_compilation.
Supplementary Material
Acknowledgments
We thank Wendy Gilbert for helpful comments during the 2019 RNA Society annual meeting. We also thank Vincent Vanoosthuyse, Christelle Morris, Geneviève Fourel, and RMI2 laboratory members for manuscript proofreading. We gratefully acknowledge support from the Pôle Scientifique de Modélisation Numérique (PSMN) of the ENS de Lyon for the computing resources. We thank the members of the LBMC biocomputing hub for their involvement in this project. We acknowledge the contribution of SFR Biosciences (UMS3444/CNRS, US8/Inserm, ENS de Lyon, UCBL) ANIRA and PBES animal facility. This work was funded by the European Research Council (ERC-StG-LS6-805500 to E.P.R.) under the European Union's Horizon 2020 research and innovation programs, by the ATIP-Avenir program (to E.P.R.), by Fondation FINOVI (to E.P.R.), and by the Howard Hughes Medical Institute (to M.J.M).
Author contributions: B.C.M., M.J.M., and E.P.R. conceived the study and designed all experiments. B.C.M., E.P.R., and A.B. performed experiments, with technical assistance from L.G.; E.L. and D.C. performed most of the bioinformatic analyses with help from A.C. for sequencing processing of sequencing reads and codon usage calculation, D.C. for the binning plot strategy and transcript database creation, F.A. for G-quadruplex analysis, L.M. for statistical analysis of generated data, and K.J. for data normalization analysis and microRNA analyses not presented in the manuscript. E.P.R. wrote the manuscript with contributions from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277863.123.
Freely available online through the Genome Research Open Access option.
Competing interest statement
M.J.M. and A.B. are employees of Moderna. All other authors declare no competing interests.
References
- Agarwal V, Kelley DR. 2022. The genetic and biochemical determinants of mRNA degradation rates in mammals. Genome Biol 23: 245. 10.1186/s13059-022-02811-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat AB, Liechti A, Matos MD, Dreos R, Janich P, Gatfield D. 2020. Transcriptome-wide sites of collided ribosomes reveal principles of translational pausing. Genome Res 30: 985–999. 10.1101/gr.257741.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar-Patel A, Kaymaz Y, Rajakumar A, Bailey JA, Karumanchi SA, Moore MJ. 2017. FLT1 and transcriptome-wide polyadenylation site (PAS) analysis in preeclampsia. Sci Rep 7: 12139. 10.1038/s41598-017-11639-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H, Coller J. 2022. Codon optimality-mediated mRNA degradation: linking translational elongation to mRNA stability. Mol Cell 82: 1467–1476. 10.1016/j.molcel.2022.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35: 2087–2103. 10.15252/embj.201694699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin J-D, Jodoin R, Perreault J-P. 2014. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res 42: 1209–1223. 10.1093/nar/gkt904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Statist 29: 1165–1188. 10.1214/aos/1013699998 [DOI] [Google Scholar]
- Bicknell AA, Ricci EP. 2017. When mRNA translation meets decay. Biochem Soc Trans 45: 339–351. 10.1042/BST20160243 [DOI] [PubMed] [Google Scholar]
- Burger K, Mühl B, Kellner M, Rohrmoser M, Gruber-Eber A, Windhager L, Friedel CC, Dölken L, Eick D. 2013. 4-Thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol 10: 1623–1630. 10.4161/rna.26214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke PC, Park H, Subramaniam AR. 2022. A nascent peptide code for translational control of mRNA stability in human cells. Nat Commun 13: 6829. 10.1038/s41467-022-34664-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschauer R, Matsuo Y, Sugiyama T, Chen Y-H, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R, et al. 2020. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368: eaay6912. 10.1126/science.aay6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyi-Ying C, Ann-Bin S. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470. 10.1016/S0968-0004(00)89102-1 [DOI] [PubMed] [Google Scholar]
- Cottalorda A, Verschelde C, Marçais A, Tomkowiak M, Musette P, Uematsu S, Akira S, Marvel J, Bonnefoy-Berard N. 2006. TLR2 engagement on CD8 T cells lowers the threshold for optimal antigen-induced T cell activation. Eur J Immunol 36: 1684–1693. 10.1002/eji.200636181 [DOI] [PubMed] [Google Scholar]
- Da Huang W, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acid Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Huang W, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Dave P, Roth G, Griesbach E, Mateju D, Hochstoeger T, Chao JA. 2023. Single-molecule imaging reveals translation-dependent destabilization of mRNAs. Mol Cell 83: 589–606.e6. 10.1016/j.molcel.2023.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois MF, Nguyen VT, Bellier S, Bensaude O. 1994. Inhibitors of transcription such as 5,6-dichloro-l-β-D-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J Biol Chem 269: 13331–13336. 10.1016/S0021-9258(17)36837-0 [DOI] [PubMed] [Google Scholar]
- Fanourgakis G, Lesche M, Akpinar M, Dahl A, Jessberger R. 2016. Chromatoid body protein TDRD6 supports long 3′ UTR triggered nonsense mediated mRNA decay. PLoS Genet 12: e1005857. 10.1371/journal.pgen.1005857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest ME, Pinkard O, Martin S, Sweet TJ, Hanson G, Coller J. 2020. Codon and amino acid content are associated with mRNA stability in mammalian cells. PLoS One 15: e0228730. 10.1371/journal.pone.0228730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, et al. 2017. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8: 16056. 10.1038/ncomms16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen SL, Giacomelli C, Hodge K, Zanivan S, Bushell M, Wilczynska A. 2021. Differential regulation of mRNA fate by the human Ccr4-Not complex is driven by coding sequence composition and mRNA localization. Genome Biol 22: 284. 10.1186/s13059-021-02494-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Müller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W, et al. 2014. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun 5: 5465. 10.1038/ncomms6465 [DOI] [PubMed] [Google Scholar]
- Hanson G, Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19: 20–30. 10.1038/nrm.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PC, Wei J, Dou X, Harada BT, Zhang Z, Ge R, Liu C, Zhang L-S, Yu X, Wang S, et al. 2023. Exon architecture controls mRNA m6A suppression and gene expression. Science 379: 677–682. 10.1126/science.abj9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer EE, Ozadam H, Ricci EP, Cenik C, Moore MJ. 2015. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acid Res 43: e2. 10.1093/nar/gku1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hia F, Yang SF, Shichino Y, Yoshinaga M, Murakawa Y, Vandenbon A, Fukao A, Fujiwara T, Landthaler M, Natsume T, et al. 2019. Codon bias confers stability to human mRNAs. EMBO Rep 20: e48220. 10.15252/embr.201948220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. 10.1016/j.cell.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–229. 10.1038/nature08265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SS, Lim J, Yu Z, Kong P, Sefik E, Xu H, Harman CCD, Kim LK, Lee GR, Li H-B, et al. 2020. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 367: 1255–1260. 10.1126/science.aax0194 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Maragkakis M, Alexiou P, Mourelatos Z. 2018. Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat Struct Mol Biol 25: 302–310. 10.1038/s41594-018-0042-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS. 2018. ZNF598 is a quality control sensor of collided ribosomes. Mol Cell 72: 469–481.e7. 10.1016/j.molcel.2018.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karousis ED, Gypas F, Zavolan M, Mühlemann O. 2021. Nanopore sequencing reveals endogenous NMD-targeted isoforms in human cells. Genome Biol 22: 223. 10.1186/s13059-021-02439-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3´UTRs so as to elicit mRNA decay. Cell 120: 195–208. 10.1016/j.cell.2004.11.050 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2013. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci USA 110: 3357–3362. 10.1073/pnas.1219908110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Choe J, Park OH, Kim YK. 2020. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet 36: 177–188. 10.1016/j.tig.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Li H-B, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al. 2017. M6a mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548: 338–342. 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Gasic I, Chandrasekaran V, Peters N, Shao S, Mitchison TJ, Hegde RS. 2020. TTC5 mediates autoregulation of tubulin via mRNA degradation. Science 367: 100–104. 10.1126/science.aaz4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X, Li H, Miao S, Chen H, He X, et al. 2022. tRNA-m1A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol 23: 1433–1444. 10.1038/s41590-022-01301-3 [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6: 26. 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, Link K, Khatwani N, Reynders J, Moore MJ, et al. 2019. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci USA 116: 24075–24083. 10.1073/pnas.1908052116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Muñoz SG, Kushawah G, Castellano LA, Diez M, DeVore ML, Salazar MJB, Bazzini AA. 2021. Crosstalk between codon optimality and cis-regulatory elements dictates mRNA stability. Genome Biol 22: 14. 10.1186/s13059-020-02251-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD. 2019. m6A-mediated translation regulation. Biochim Biophys Acta Gene Regul Mech 1862: 301–309. 10.1016/j.bbagrm.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels H-H, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, et al. 2015. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161: 1058–1073. 10.1016/j.cell.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Tomari Y. 2016. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61: 874–885. 10.1016/j.molcel.2016.02.027 [DOI] [PubMed] [Google Scholar]
- Morris C, Cluet D, Ricci EP. 2021. Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny. Wiley Interdiscip Rev RNA 12: e1658. 10.1002/wrna.1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula A, Ellis J, Taliaferro JM, Rissland OS. 2019. Coding regions affect mRNA stability in human cells. RNA 25: 1751–1764. 10.1261/rna.073239.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasif S, Contu L, Mühlemann O. 2018. Beyond quality control: the role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin Cell Dev Biol 75: 78–87. 10.1016/j.semcdb.2017.08.053 [DOI] [PubMed] [Google Scholar]
- Nicolet BP, Zandhuis ND, Lattanzio VM, Wolkers MC. 2021. Sequence determinants as key regulators in gene expression of T cells. Immunol Rev 304: 10–29. 10.1111/imr.13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Seo J-W, Ahn N, Park S, Hwang J, Nam J-W. 2019. UPF1/SMG7-dependent microRNA-mediated gene regulation. Nat Commun 10: 4181. 10.1038/s41467-019-12123-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2015. Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161: 1400–1412. 10.1016/j.cell.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen Y-H, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen Y-H, Martin S, Alhusaini N, Green R, Coller J. 2016. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167: 122–132.e9. 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak R, Polonsky M, Eizenberg-Magar I, Mo Y, Sakaguchi Y, Mizrahi O, Nachshon A, Reich-Zeliger S, Stern-Ginossar N, Dahan O, et al. 2021. Dynamic changes in tRNA modifications and abundance during T cell activation. Proc Natl Acad Sci USA 118: e2106556118. 10.1073/pnas.2106556118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2023. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 21: 26–35. 10.1038/nsmb.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Pérez R, Bajorath J. 2020. Interpretation of machine learning models using shapley values: application to compound potency and multi-target activity predictions. J Comput Aided Mol Des 34: 1013–1026. 10.1007/s10822-020-00314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H, Donnard E, Liu B, Jung S, Wang R, Richter JD. 2020. FMRP links optimal codons to mRNA stability in neurons. Proc Natl Acad Sci USA 117: 30400–30411. 10.1073/pnas.2009161117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, Barna M. 2017. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169: 1051–1065.e18. 10.1016/j.cell.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Dikstein R. 2020. So close, no matter how far: multiple paths connecting transcription to mRNA translation in eukaryotes. EMBO Rep 21: e50799. 10.15252/embr.202050799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Burge CB, Bartel DP. 2013. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 23: 2078–2090. 10.1101/gr.156919.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. 2017. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell 65: 751–760.e4. 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesina P, Heckel E, Cheng J, Fromont-Racine M, Buschauer R, Kater L, Beatrix B, Berninghausen O, Jacquier A, Becker T, et al. 2019. Structure of the 80S ribosome–Xrn1 nuclease complex. Nat Struct Mol Biol 26: 275–280. 10.1038/s41594-019-0202-5 [DOI] [PubMed] [Google Scholar]
- Timmers H, Tora L. 2018. Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol Cell 72: 10–17. 10.1016/j.molcel.2018.08.023 [DOI] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He Q-L, Bhat S, Low W-K, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. 2011. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 7: 182–188. 10.1038/nchembio.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta E. 2014. On the normalization of the minimum free energy of RNAs by sequence length. PLoS One 9: e113380. 10.1371/journal.pone.0113380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Rankova A, Arpat AB, Liechti LA, Hess D, Iesmantavicius V, Castelo-Szekely V, Gatfield D, Bühler M. 2020. Mammalian RNA decay pathways are highly specialized and widely linked to translation. Mol Cell 77: 1222–1236.e13. 10.1016/j.molcel.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzonyi A, Dierks D, Nir R, Kwon OS, Toth U, Barbosa I, Burel C, Brandis A, Rossmanith W, Le Hir H, et al. 2023. Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol Cell 83: 237–251.e7. 10.1016/j.molcel.2022.12.026 [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SEW, Nerlov C, Porse BT. 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22: 1381–1396. 10.1101/gad.468808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M, Bazzini AA. 2019. Translation affects mRNA stability in a codon dependent manner in human cells. eLife 8: e45396. 10.7554/eLife.45396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Xiong M, Xu C, Xiang P, Zhong X. 2016. Long noncoding RNAs: an overview. In Long non-coding RNAs: methods and protocols (ed. Feng Y, Zhang L), pp. 287–295. Springer, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.