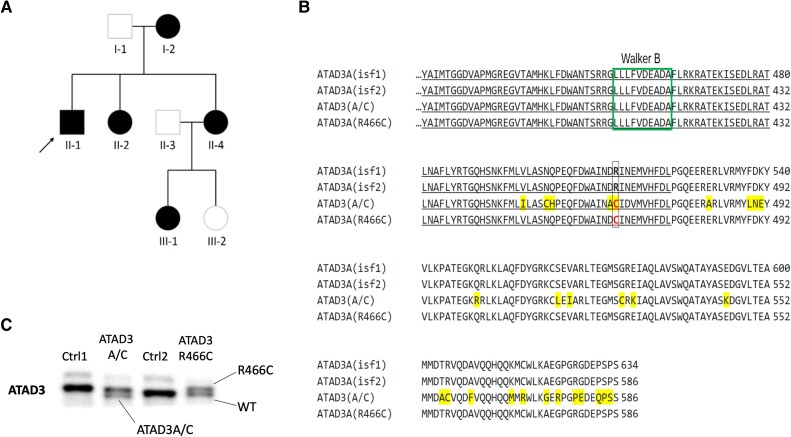
Figure 1.
Identification of a family with DOA+ associated with a dominant point mutation in ATAD3A. (A) Pedigree of the family in which affected members carry ATAD3A c.1396C>T, p.R466C (NM_001170535.2) on one allele. (B) Amino acid sequence alignment between parts of ATAD3A isoforms 1 and 2 affected by the ATAD3A/C gene fusion7 and the arginine finger point mutation. The A/C fusion protein differs at 29 amino acid positions (highlighted in yellow).6 The individuals with the R466C point mutation have an ATAD3 sequence identical in length to ATAD3A isoform 2 and it only differs in the ATP-binding residue in position 466, which it shares with the fusion protein A-C (in red font). Underlined are the residues of the conserved protein kinase domain that form the ATPase region [p.Ile348–p.Asp474; PFam PF00004]. Marked in a green box is the Walker B ATP binding motif. Residue numbering from [Q9NVI7-2/NM_001170535.2]. (C) SDS-PAGE (8% gel) of whole cell protein from controls (Ctrl), ATAD3 R466C and ATAD3A/C mutant cell lines, immunolabelled with an ATAD3 N-terminal antibody. DOA+ = dominant optic atrophy ‘plus’.