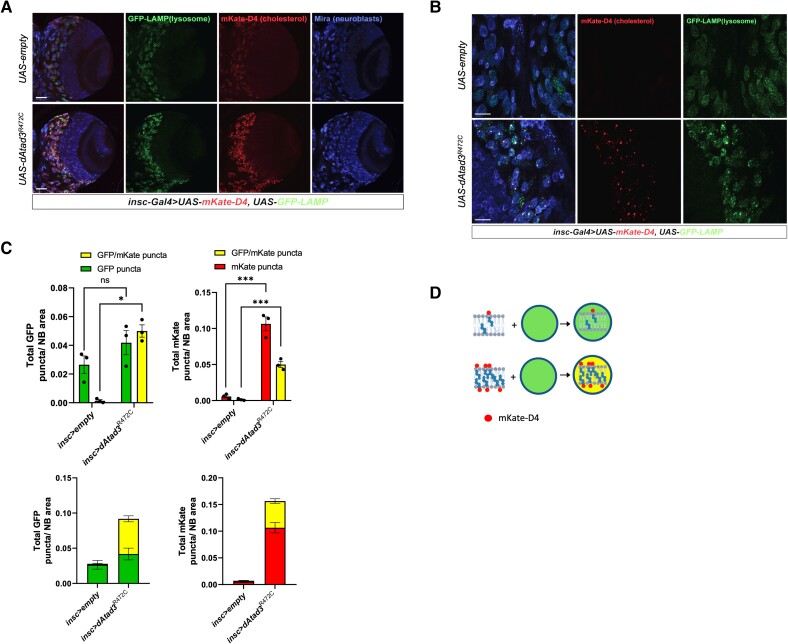
Figure 5.
ATAD3A mutant neuroblasts show increased membrane-associated cholesterol together with increased lysosome content. (A and B) Confocal micrographs of Drosophila larval brain lobes carrying insc-Gal4 (neuroblast driver), UAS-GFP-LAMP and UAS-mKate-D4 together with UAS-empty (control), or UAS-dAtad3R472C at magnifications of ×20 (scale bar = 50 µm) and ×63 (scale bar = 20 µm), respectively. Neuroblasts are labelled Miranda (blue), GFP-LAMP (green) marks lysosomes, while mKate-D4 (red) marks cholesterol-containing membranes. (C) Top left: Quantification of GFP-LAMP puncta without (green) and with mKate-D4 (yellow) per neuroblast (NB) area, for UAS-empty (control) and dAtad3R472C expressed under neuroblast driver insc-Gal4. Differences between groups were analysed by unpaired, two-tailed Student’s t-test. Green fluorescent protein (GFP) dAtad3 versus GFP empty non-significant, P = 0.2149; GFP/mKate empty versus GFP/mKate dAtad3, *P = 0.0353. Below the total number of lysosomes in a single column (green + yellow) for UAS-empty (control) and dAtad3R472C. Top right: Quantification of mKate-D4 puncta without (red) and with GFP-LAMP (yellow) per neuroblast (NB) area, for UAS-empty (control) and dAtad3R472C expressed under neuroblast driver insc-Gal4. Differences between groups were analysed by unpaired, two-tailed Student’s t-test: mKate empty versus mKate dAtad3, ***P = 0.0005; GFP/mKate empty versus GFP/mKate dAtad3, ***P = 0.0004. (D) Schematic representation of the data: cholesterol in membranes is labelled red by mKate-D4, while GFP-LAMP stains lysosomes green; hence, mKate-D4 foci and lysosomes that co-localize appear yellow.