Fig. 6 |. JP1302 blocks transcription through disruption of the FACT complex and histone H1.
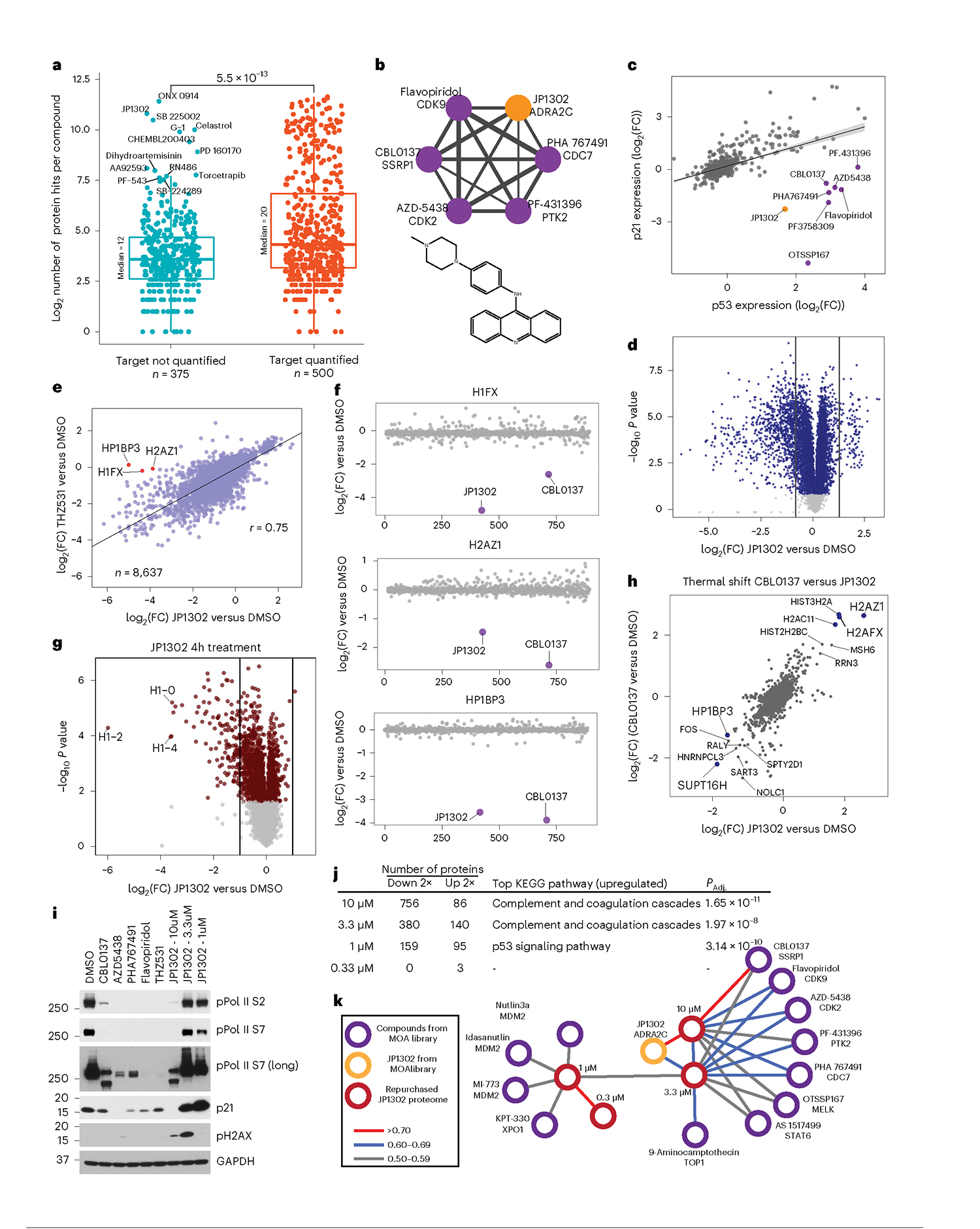
a, Number of regulated events for each compound in this dataset, based on whether a target was identified. All 2,485 annotated targets were used. The P value is from unpaired, Wilcoxon test (n = 875). The center line represents the median, the upper and lower bounds of the box indicate the IQR (range between the 25th and 75th percentiles) and whiskers extend to the highest and lowest values within 1.5 times the IQR. b, Compound–compound community plot and structure of JP1302. Edge thickness denotes the degree of correlation (r = 0.6 to 0.8). c, Protein-level regression of p53 and p21 for all compounds in the dataset. Shading along the regression line represents the 95% confidence interval. d, Volcano plot of JP1302-treated (10 μM, 24 h) cells using repurchased compound. e, Correlation of proteome fingerprints from JP1302-treated (10 μM) cells versus THZ531-treated (1 μM) cells. f, Expression of proteins highlighted in e across the MOA dataset. g, Volcano plot of protein expression from HCT116 cells treated with JP1302 (10 μM, 4 h). h, Correlation of ligand-induced changes in proteome thermal stability for CBL0137 and JP1302-treated HCT116 cells. i, Western blot of various RNA pol II and DNA damage markers in HCT116 cells treated for 20 h with indicated compounds. Treatment concentration is 10 μM or 1 μM (THZ531) unless indicated. j, Table showing the number of up- and downregulated proteins (by twofold versus DMSO) in cells treated with the indicated concentrations of JP1302. KEGG pathways enriched from the list of proteins upregulated by twofold is shown for each concentration. Enriched pathways and their adjusted (using Benjamini–Hochberg) P values (PAdj.) were calculated using the enrichKEGG R package. k, Compound–compound community plot generated from the larger MOA dataset after including the JP1302 dose-dependent proteome fingerprints (red nodes). All JP1302 nodes and all network edges higher than 0.5 (Pearson r) are shown. P values in d and g are calculated as described in Methods. Measurements with Q > 0.05 are light gray.
