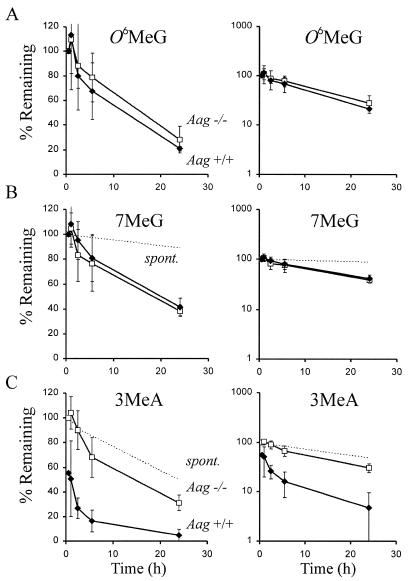
Figure 3.
Graphs on the left and right are identical, except for the change in scale of the y-axis. Percent of alkylated bases remaining in the genome of Aag–/– (opened squares) and Aag+/+ (closed diamonds) cells relative to t = 0.5 h. Spontaneous depurination of 7MeG and 3MeA at 37°C in PBS are shown as dotted lines that intercept the y-axis at 100% (values for spontaneous depurination of 3MeA were measured as described in Materials and Methods; values for 7MeG were calculated based on published estimates of t1/2; 6,36). The adduct levels at 24 h have been adjusted to correct for dilution due to DNA replication. These data are the average of four or five experiments using AB1 Aag+/+ cells and four independent Aag–/– clones. (A) O6MeG and (B) 7MeG removal from the DNA of Aag–/– and Aag+/+ cells. (C) 3MeA removal from the DNA of Aag+/+ cells and Aag–/– cells. Note that the values plotted for percent 3MeA remaining in Aag+/+ cells are the observed values divided by 1.8, to reflect the 1.8-fold difference in initial 3MeA levels between Aag+/+ and Aag–/– cells (shown in Fig. 2).