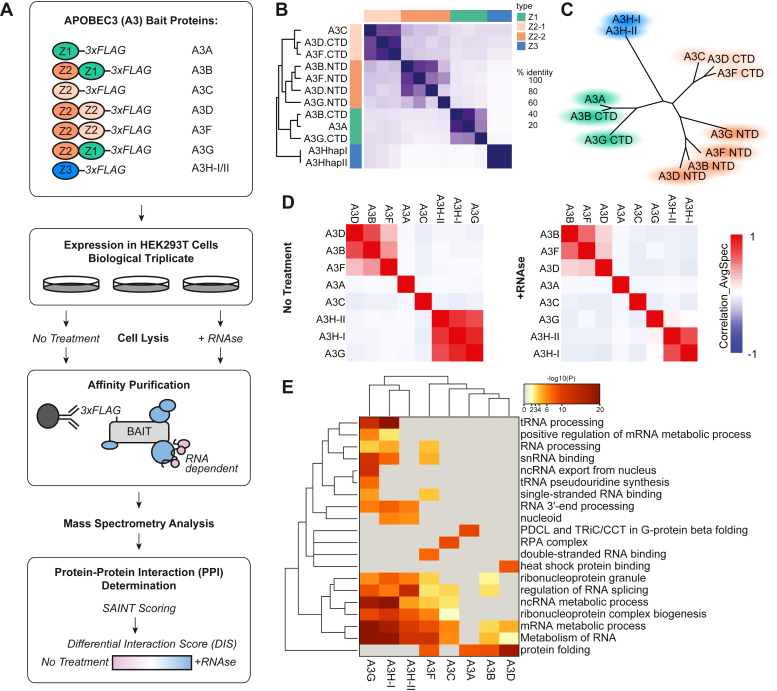
Fig. 1.
AP-MS analysis of the A3 family proteins identifies shared and specific RNA-dependent and -independent PPIs.A, in the AP-MS workflow described in this study, HEK293T cells were transiently transfected with C-terminally 3× FLAG-tagged A3 proteins in biological triplicate and purified with or without RNAse A treatment. Purified proteins were trypsin digested and analyzed by LC-MS/MS. Peptides and proteins were identified by MaxQuant (28), high confidence PPIs were determined by SAINT and CompPASS, and RNA-dependent and independent PPIs were characterized using a Differential Interaction Score (DIS) calculation. The cartoon representation at the top shows each of the 3× FLAG tagged A3 proteins with the Z1 domain in green, the Z2 domain in orange, and Z3 domain in blue. B, sequence identity matrix demonstrating pairwise similarity in A3 domains. C, clustered tree diagram of the similarity of A3 protein domains. D, correlation plot of SAINT-scored proteins identified in A3 pull-downs with no treatment (left) and RNAse A treatment (right); performed by Prohits-viz (40). E, heatmap of the top functional enrichment analysis of high confidence PPIs per each bait. For the full list of enrichments see Supplemental Table S7.