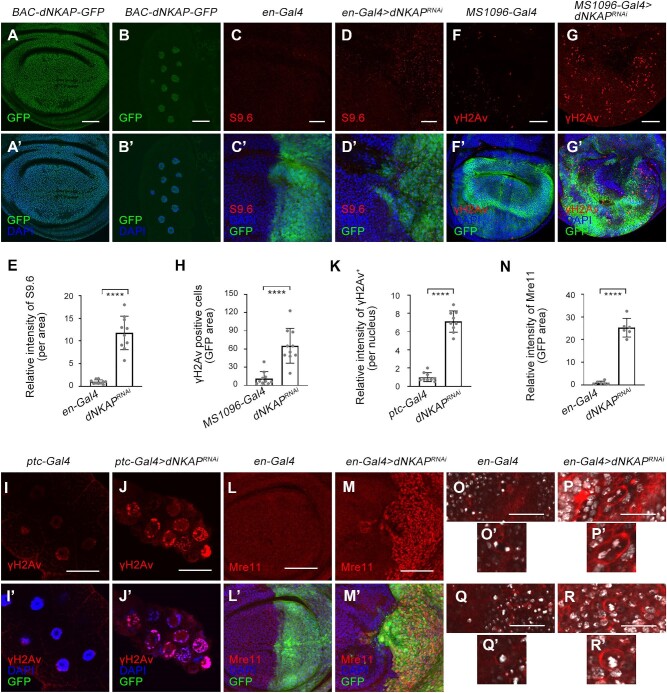
Figure 2.
dNKAP localizes to the nucleus and functions to maintain genome stability. (A–B’) Nuclear localization of dNKAP in mitotic wing disc (A and A’) and polyploid salivary gland (B and B’) cells. Scale bar, 50 μm. (C–D’) Accumulation of R-loops in dNKAP-deficient wing disc cells. Scale bar, 20 μm. (E) Quantification of S9.6 signals from C and D. n = 10. (F–G’) Increased γH2Av signal in dNKAP-deficient wing disc cells. Scale bar, 50 μm. (H) Quantification of γH2Av signals from F and G. n = 10. (I–J’) Increased γH2Av signal in dNKAP-deficient salivary gland cells. Scale bar, 50 μm. (K) Quantification of γH2Av signals from I and J. n = 9. (L–M’) Increased Mre11 protein level in dNKAP-deficient wing disc cells. Scale bar, 50 μm. (N) Quantification of Mre11 signals from L and M. n = 6. Note that three different regions with the same size were calculated per disc. (O–R’) Chromosome abnormalities in dNKAP-deficient wing disc cells. Note the chromosome bridge and micronucleus phenotype in dNKAP-deficient cells. Scale bar, 150 μm. GFP was used to mark the knockdown domain. Panels C–D’ and I–J’ show single confocal sections of third-instar wing imaginal discs with posterior side to the right and dorsal side up. Panels F and G show the maximum projections across image slices. ****P < 0.0001.