Figure 4.
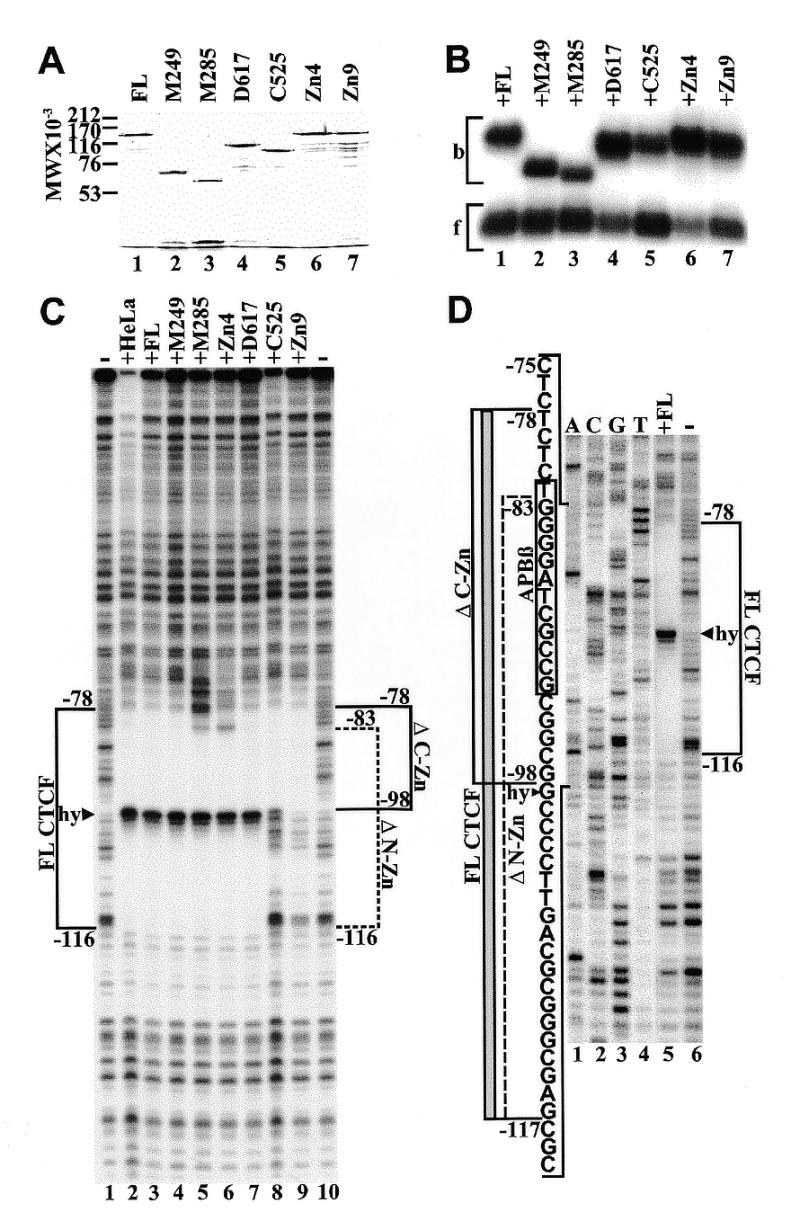
Binding of purified recombinant CTCF constructs to APP[WT]. (A) Coomassie blue stained SDS–polyacrylamide gel of full-length CTCF (lane 1), N-terminal deletions M249 (lane 2), M285 (lane 3), C-terminal deletions D617 (lane 4), C525 (lane 5), and internal zinc finger deletions Zn4 (lane 6) and Zn9 (lane 7). (B) Mobility shift electrophoresis of the same CTCF constructs shown in (A). All CTCF constructs were bound to radiolabeled oligonucleotide APBβ[WT] (Fig. 1C). Bound (b) and free (f) oligoncleotides are indicated by brackets. (C) DNase I footprinting of purified CTCF constructs bound to 5′-end-labeled wild-type APP fragment extending from position –193 to +100. Fragments were 5′-end-labeled at position –193 and digested with DNase I either in the absence of CTCF (lanes 1 and 10), or with full-length CTCF purified from HeLa cell nuclear extract (lane 2), full-length recombinant CTCF from P.pastoris (lane 3), N-terminal deletions M249 (lane 4) and M285 (lane 5), internal zinc finger deletion Zn4 (lane 6), C-terminal deletions D617 (lane 7) and C525 (lane 8), and internal zinc finger deletion Zn9 (lane 9). The DNase I footprint of full-length CTCF (FL CTCF) is delineated by a bracket from position –78 to –116 interrupted by a hypersensitive site (hy, arrowhead). Brackets also indicate the DNase I footprints generated by N-terminal (ΔN-Zn, position –83 to –116) and C-terminal (ΔC-Zn, position –98 to –78) zinc finger deletions. (D) DNase I footprinting of 5′-end-labeled wild-type APP fragment without CTCF (lane 1) or with bound full-length CTCF (lane 2). The DNase I footprinting reactions were co-electrophoresed with a dideoxy sequencing reaction obtained with the same 32P-labeled oligonucletide used for PCR amplification (lanes 3–6). The sequence within the bracketed region is provided together with the outlines of DNase I protected domains obtained with FL CTCF (gray bar). Brackets also indicate the outlines of the DNase I protected domains obtained with N- (ΔN-Zn) and C-terminal (ΔC-Zn) deletions of CTCF. The sequence of the APBβ binding sequence is boxed. The boundaries of the footprints are defined by the terminal nucleotides protected from DNase I digestion and their position should be considered accurate within 1 bp.
