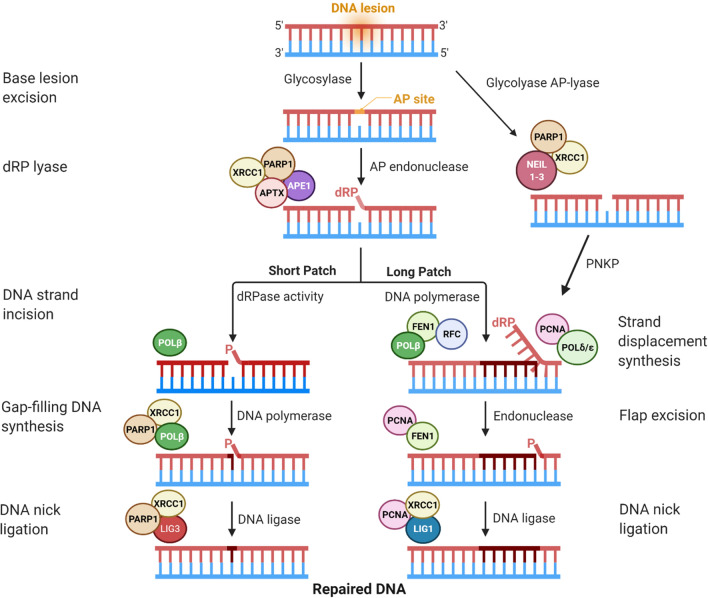
Fig. 3.
A simplified scheme illustrates the Short Patch and Long Patch base excision repair (BER) pathways in eukaryotes. Lesion-specific DNA glycosylases (e.g., UNG1) recognize and remove the damaged base resulting in an abasic site. An APE1 incision follows this to create a single-strand break with 3′-hydroxyl and 5′-dRP ends. The POLβ-dRP lyase activity excises the latter, and POLβ simultaneously fills the gap with either single-nucleotide (left BER, Short Patch) or 2 to 11 nucleotides with FEN1/RFC coupling (right BER, Long Patch). The choice between Short Patch BER or Long Patch BER depends on the state of the 5′dRP end. NEIL-DNA glycosylases (NEIL1-3) contain a β,δ-elimination activity that results in a single-nucleotide gap with a 3′-phosphate end. PNKP will then remove the 3′-phosphate, and the pathway may proceed via Long Patch Repair. Finally, ligation of the DNA-strand nicks is performed through a LIG3-XRCC1-PARP1 complex to complete the Short Patch BER. The polymerase activity will be switched to POLδ/ε when the 5′-dRP termini resist POLβ activity, which can add 2 to 11 nucleotides in the gap. This pathway leaves a flap recognized and removed via FEN1 endonuclease activity that forms a complex with PCNA (right branch). The Long Patch BER is completed when the remaining DNA backbone nick is sealed by DNA LIGI, also associated with PCNA and XRCC1. Orange haze depicts the modified base, and nascent nucleotide(s) are shown in dark red. AP site apurinic/apyrimidinic site, APE1 AP-endonuclease 1, dRP 5′-deoxyribose phosphate, POL DNA polymerase, XRCC1 X-ray cross-complementing protein 1, LIG1-3 DNA ligase I and 3, PCNA proliferating cell nuclear antigen, RFC replication factor C, FEN1 Flap endonuclease, PARP1 poly (ADP-ribose) polymerase, PNKP polynucleotide kinase phosphatase, APTX aprataxin, NEIL1-3 endonuclease VIII-like glycosylases 1, 2, and 3. The figure was generated with a license from BioRender.com