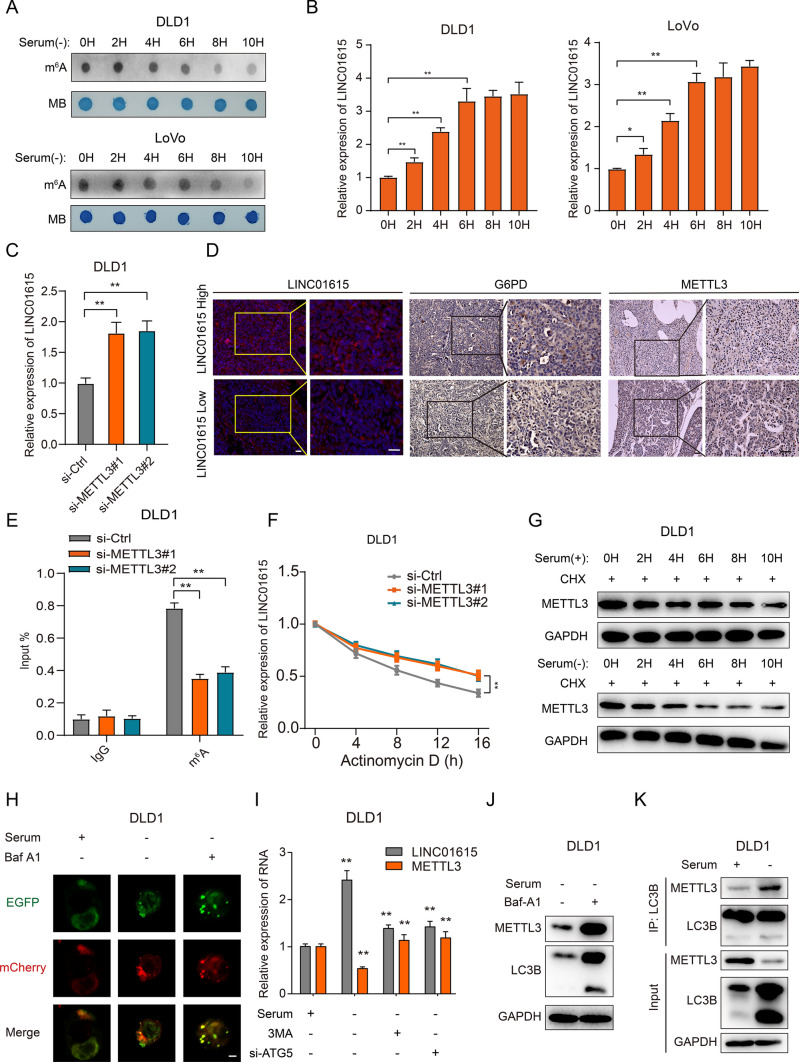
Fig. 6.
Autophagy-reduced METTL3 upregulates LINC01615 in a m6A modification-dependent manner. A A dot blot assay was used to analyze the m6A content of total RNA in DLD1 and LoVo cells cultured without serum supplementation for different times. Methylene blue staining was used as a loading control. B qRT–PCR analysis of LINC01615 expression in DLD1 and LoVo cells cultured without serum supplementation for different times. C qRT–PCR analysis of LINC01615 expression in DLD1 cells transfected with METTL3 siRNA or control siRNA. D Representative images of LINC01615, METTL3 and G6PD analyzed by FISH or IHC in LINC01615 high-expression and LINC01615 low-expression tumors. E MeRIP-qPCR showing the m6A modification enrichment of LINC01615 in DLD1 cells after METTL3 depletion. F qRT–PCR analysis of the stability of LINC01615 in DLD1 cells with or without METTL3 deletion. G WB analysis of METTL3 protein stability in DLD1 cells cultured with or without serum supplementation for different times. H Representative confocal microscopy images of LC3B puncta distribution in DLD1 cells under normal or serum-free starvation conditions and treated with or without Baf-A1 (1000 × magnification). I Analysis of the expression of METTL3 and LINC01615 in DLD1 cells transfected with ATG5 siRNA, METTL3 overexpression plasmid, control vector or treated with 3-MA 30 min before the end of each experimental time. J WB analysis of METTL3 expression in DLD1 cells treated with or without Baf-A1 under serum-free starvation conditions. K Co-IP analysis was performed with specific antibodies against METTL3 and LC3B in DLD1 cells under normal or serum-free starvation conditions. WB analysis of the interaction between METTL3 and LC3B. Data are the means ± SDs (n = 3 independent experiments), *p < 0.05, **p < 0.01