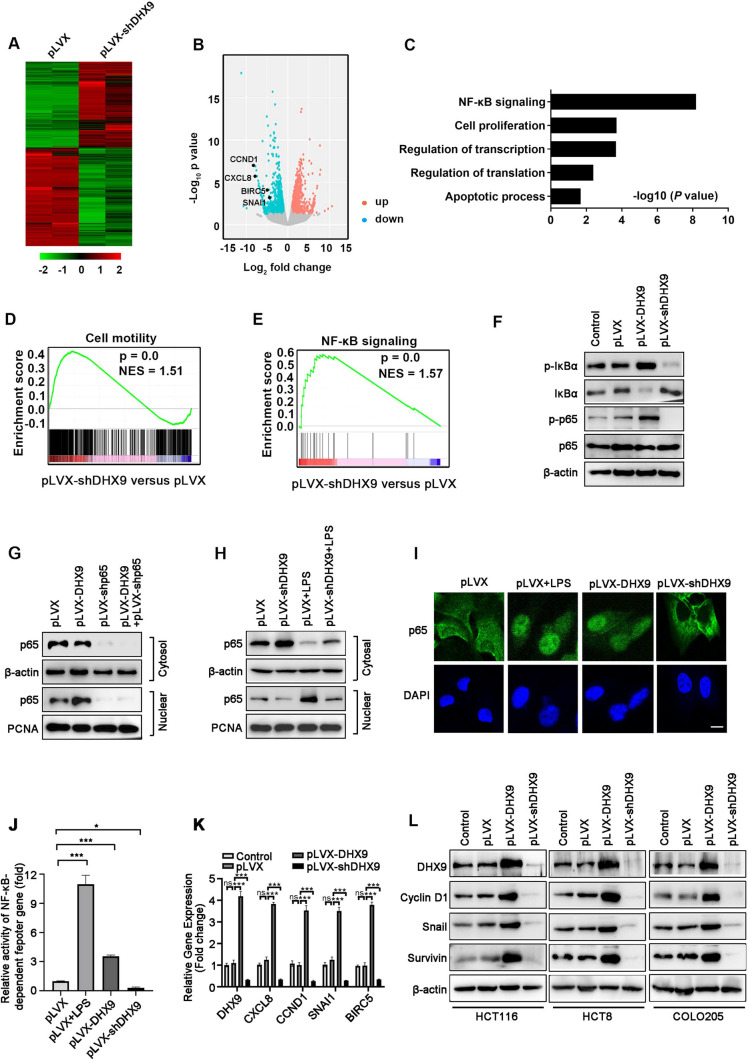
Fig. 6.
DHX9 mediates the activation of NF-κB signal pathway in colorectal cancer cells. A Heatmap showed the alteration of gene expression (≥ twofold) obtained from RNA-Seq of vector-transfected and DHX9-silenced HCT116 cells. B Scatter plot displayed fold changes of gene expression (≥ twofold) in DHX9-silenced HCT116 cells compared with vector-transfected cells. C GO analysis of down-regulated genes (≥ twofold) in DHX9-silenced HCT116 cells compared with vector-transfected cells. D-E GSEA revealed that gene sets of cell motility D and NF-κB signaling E were enriched in DHX9-depleted HCT116 cells. F Proteins levels of IKBα, p65 and phosphorylated IKBα and p65 were evaluated by Western blotting in DHX9-overexpressed or -depleted HCT116 cells. G-H Cytosolic and nuclear fractionations were evaluated by Western blotting in DHX9-overexpressed HCT116 cells with or without p65 knockdown G or in DHX9-silenced HCT116 cells with or without LPS (1 μg/ml) pretreatment for 15 min H. I DHX9-overexpressed or -depleted HCT116 cells were subjected to immunofluorescence analysis of p65 localization (green). Vector-transfected (pLVX) HCT116 cells pretreated with LPS (1 μg/ml) for 15 min were served as a positive control. DAPI (blue) was applied to stain the nuclear. Scale bar, 20 μm. J HCT116 cells with stably expressing pLVX, pLVX-DHX9 or shRNAs against DHX9 were co-transfected with 0.5 μg NF-κB-TATA-Luc reporter plasmid and 10 ng Renilla luciferase reporter. 36 h after transfection, pLVX-HCT116 cells were treated with LPS (200 ng/mL) for 8 h. The luciferase activity of cells was measured. The values of firefly luciferase activity were normalized to Renilla luciferase activity. Fold activation were represented as mean ± SEM of three independent transfections. ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test. K RT-qPCR experiment analyzed NF-κB-target genes including CXCL8, encoding IL-8; CCND1, encoding Cyclin D1; BIRC5, encoding Survivin; SNAI1, encoding Snail, in DHX9-overexpressed or -depleted HCT116 cells. Data were from three independent experiments and represented as mean ± SEM. ns, not significant; ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test. L Western blotting analysis verified the protein levels of candidate NF-κB-target genes in DHX9-overexpressed or -depleted cells