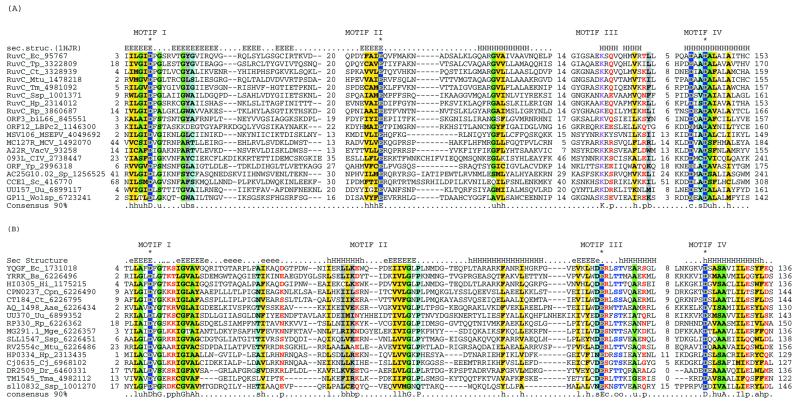
Figure 1.
Multiple alignments of the HJRs of the RNase H fold. (A) RuvC family. (B) YqgF family. Each protein is labeled using the gene name followed by the species abbreviation and the GenBank gene identifier. The extent of the domain in each protein is indicated by numbers to the sides of the alignments. The long poorly conserved inserts are replaced by numbers indicating the number of omitted residues. Only Motifs I and IV are aligned between the RuvC and YqgF families. The secondary structure predicted using the PHD program is shown above the alignment with H/h for α-helix and E/e for β-strands (upper case denoting strong prediction and lower case moderate prediction). The shading and coloring are according to the 90% consensus, which is shown underneath the alignment, with the following convention: h, hydrophobic residues (YFWLIVMA); l, aliphatic residues (LIVMA); a, aromatic residues (YFWH), yellow background; p, polar residues (STQNEDRKH), red foreground; s, small residues (SAGTVPNHD), turquoise background; u, tiny residues (GAS), light green background; c, charged residues (KHRED), magenta foreground; b, bulky residues (LIYWFEQRKM), gray background. The residues predicted to form the active site or associated with catalysis are shown in inverse coloring. The species abbreviations are as indicated in Table 1. The following are abbreviations not shown in Table 1: Wolsp, Wolbachia sp.; MSEPV, Melanopus sanguinus entomopox virus; MCV, Molluscum contagiousum virus; VacV, vaccinia virus; biL66 and LBPc2, lactococcal phages biL66 and c2.