Abstract
In human metabolism, pyruvate dehydrogenase complex (PDC) is one of the most intricate and large multimeric protein systems representing a central hub for cellular homeostasis. The worldwide used antiepileptic drug valproic acid (VPA) may potentially induce teratogenicity or a mild to severe hepatic toxicity, where the underlying mechanisms are not completely understood. This work aims to clarify the mechanisms that intersect VPA-related iatrogenic effects to PDC-associated dihydrolipoamide dehydrogenase (DLD; E3) activity. DLD is also a key enzyme of α-ketoglutarate dehydrogenase, branched-chain α-keto acid dehydrogenase, α-ketoadipate dehydrogenase, and the glycine decarboxylase complexes. The molecular effects of VPA will be reviewed underlining the data that sustain a potential interaction with DLD. The drug-associated effects on lipoic acid-related complexes activity may induce alterations on the flux of metabolites through tricarboxylic acid cycle, branched-chain amino acid oxidation, glycine metabolism and other cellular acetyl-CoA-connected reactions. The biotransformation of VPA involves its complete β-oxidation in mitochondria causing an imbalance on energy homeostasis. The drug consequences as histone deacetylase inhibitor and thus gene expression modulator have also been recognized. The mitochondrial localization of PDC is unequivocal, but its presence and function in the nucleus were also demonstrated, generating acetyl-CoA, crucial for histone acetylation. Bridging metabolism and epigenetics, this review gathers the evidence of VPA-induced interference with DLD or PDC functions, mainly in animal and cellular models, and highlights the uncharted in human. The consequences of this interaction may have significant impact either in mitochondrial or in nuclear acetyl-CoA-dependent processes.
Keywords: Dihydrolipoamide dehydrogenase, Pyruvate dehydrogenase, Protein lysine acetylation, Mitochondrial dysfunction, Valproate, Valproate hepatotoxicity, Valproate teratogenicity
Introduction
The liver is an essential organ that plays vital functions in the maintenance of metabolic homeostasis, which is crucial for the biotransformation of many drugs. Mitochondrial energy metabolism is highly regulated to satisfy the metabolic needs of the cell. The mechanisms of mitochondrial dysfunction have been the focus of our research interests, namely as the fundamental basis of drug-induced iatrogenic effects with impact on liver function. Mitochondria produce energy in the form of adenosine triphosphate (ATP) by the process of oxidative phosphorylation (OXPHOS) but other relevant processes, such as fatty acid β-oxidation (FAO), tricarboxylic acid (TCA) cycle, urea cycle, and ketone body production play a key role in mitochondrial bioenergetics [1, 2].
Pyruvate is an essential product of glycolysis that may enter the mitochondria for further oxidation. Pyruvate dehydrogenase complex (PDC) catalyzes the oxidation of pyruvate into acetyl-CoA, an irreversible reaction, connecting glycolysis to the TCA cycle [3]. This multienzyme system is composed of three distinct catalytic components: pyruvate dehydrogenase/decarboxylase (E1/PDH), dihydrolipoamide transacetylase/acetyltransferase (E2/DLAT), and dihydrolipoamide dehydrogenase (E3/DLD), as well as one tethering component, the E3-binding protein (E3BP) [4, 5]. It is important to underline that, besides PDC, the E3 protein is also part of four additional mitochondrial complexes namely, α-ketoglutarate dehydrogenase (α-KGDC), branched-chain α-keto acid dehydrogenase (BCKADC), α-ketoadipate dehydrogenase (α-KADC) and the glycine decarboxylase complex (GDC) also known as glycine cleavage system (GCS). Considering the pivotal role of DLD at prominent catalytic clusters, we postulate that any drug-associated effect on this protein of widespread designation as E3, and/or on PDC activity may induce pleiotropic effects in cells and tissues. Pyruvate oxidation deficiencies secondary to acquired causes such as medicines are far from being recognized, whereas the genetic causes represent a well-defined group of mitochondrial diseases [6]. The localization of PDC has been thought for years to be strictly mitochondrial [7]. Interestingly, its functional role in the nucleus was demonstrated, generating acetyl-Coenzyme A (CoA), a substrate for histone acetylation [8]. The presence of PDC in the nucleus links metabolism and epigenetics, a determinant process for gene expression and energy homeostasis, and seems to be associated with mitochondrial stressors [8].
Valproic acid (VPA) is a worldwide used antiepileptic drug that has been recognized as a lysine deacetylase inhibitor (KDACi) of histones [9], whose potential anti-cancer effects are under evaluation in numerous clinical studies. Beyond its well-established clinical use, this drug has been reported to induce mild to severe hepatic toxicity and possible teratogenicity, but the underlying mechanisms are not completely understood. VPA is a simple branched-chain fatty acid and its overall metabolism, activation to valproyl-CoA and respective drug effects on mitochondrial FAO have been reviewed [10]. Once inside mitochondria, and in the presence of ATP and CoA, VPA is converted to valproyl-CoA (VP-CoA), an active intermediate that undergoes complete β-oxidation in this organelle. The sequential enzyme-mediated reactions of the cycle up to thiolytic cleavage, will produce propionyl-CoA and acetyl-CoA, that ultimately enter the TCA cycle to complete oxidation to CO2 and H2O. This metabolic pathway typically accounts for ca. 40% of the overall metabolism of the drug. Various targeted effects on mitochondrial processes were demonstrated including a VPA-induced decrease on FAO as an underlying cause of hepatic steatosis [10]. However, at the molecular level other consequences are noteworthy, where a clear inhibition of pyruvate oxidation in liver mitochondria has been reported, which may trigger undisputed effects on energy metabolism [11].
The present review focuses on the molecular effects of this drug as a potential inhibitor of DLD (E3) or overall PDC with a clear impact on the flux of metabolites through TCA cycle and other acetyl-CoA dependent cellular reactions. The understanding of the metabolic bases by which VPA or valproyl-CoA may interfere with all DLD-dependent complexes in cells is incomplete. Highlighting the function and intracellular localization of proteins or metabolites, this work will gather insights into the multiple mechanisms of drug-induced liver toxicity, where interference with E3 or PDC function will certainly play a pivotal role.
Pyruvate mitochondrial metabolism
Pyruvate is a key metabolite that intersects crucial pathways of energy metabolism. In the cytosol, pyruvate is produced from glucose through glycolysis, or is generated by transamination of alanine. It may also represent a precursor for gluconeogenesis, synthesis of glycerol, fatty acids, and some amino acids, and thus regulatory mechanisms of the fate of pyruvate are fundamental for homeostasis. As a polar anion (conjugate base of the α-ketoacid, pyruvic acid), it does not easily penetrate nor freely diffuses across subcellular membranes. To enter mitochondria, pyruvate needs to cross the outer mitochondrial membrane probably through the large, voltage-dependent anion channel (VDAC), attaining the intermembrane space. It is then transported, together with a proton, across the inner mitochondrial membrane through the mitochondrial pyruvate carrier (MPC), into the matrix. The existence of MPC has been studied for decades, but its molecular characterization was only achieved in 2012 [12, 13]. In most mammalian cells, the active MPC is an obligate heterodimer that comprises two small hydrophobic integral membrane subunits, MPC1 and MPC2, essential and necessary for pyruvate transport into the mitochondria. Thus, MPC serves as a unique gatekeeper for pyruvate entry into mitochondria linking glycolysis with mitochondrial respiration and OXPHOS. The role of MPC in health or in disease [14], such as cancer or tumorigenesis [15, 16], is undisputed. It regulates the function of many organs in human, including the heart or brain [17–19].
Pyruvate is a main entry intermediate to the TCA cycle. Once inside mitochondria, two enzymes may direct pyruvate to two major and distinct reactions: i) carboxylation via pyruvate carboxylase (PC), the enzyme that catalyzes the ATP-dependent conversion of pyruvate to oxaloacetate (OAA) or ii) oxidation via PDC, the enzymatic system that mediates acetyl-CoA formation. The flux through the TCA cycle starts with the transfer of the two-carbon acetyl group from acetyl-CoA to OAA, the four-carbon acceptor compound, to form a six-carbon compound (citrate). This condensation is irreversible and catalyzed by citrate synthase representing the first committed step of TCA cycle. Considering the initial reactions of pyruvate, the role of PC mediating OAA formation represents a key anaplerotic pathway that contributes to replenish the intermediates of TCA cycle [20]. PC catalyzes, in a tissue-specific manner, the initial reactions of glucose (liver, kidney) and lipid (adipose tissue, liver, brain) synthesis from pyruvate. While irreversible decarboxylation of pyruvate catalyzed by PDC yielding acetyl-CoA occurs in mitochondria of all cell types, PC activity shows a greater cell-specific distribution with higher expression in liver and adipose tissue and much lower in the brain neurons.
Since cells contain mitochondrial and cytosolic isoforms of alanine transaminase (ALT), pyruvate can be converted to alanine in the cytosol, which enters the mitochondria via an alanine carrier and is transaminated back to pyruvate inside the mitochondrial matrix for further metabolism by PDC or PC [14, 21]. Interestingly, a study using liver-specific MPC2 knockout mice demonstrated a pyruvate-alanine cycling mechanism to avoid potential loss of MPC function, to maintain gluconeogenic flux and hepatic glucose homeostasis [22].
PDC and major regulatory enzymes
PDC and its regulatory enzymes play a crucial role in cellular metabolism in all mitochondria of higher eukaryotes. This complex is active in aerobic conditions that require glucose decomposition to produce energy but is suppressed when glucose is limited [3–5]. PDC belongs to the family of often called 2-oxo or α-ketoacid dehydrogenase complexes, which are composed of several and different interacting enzymes that are connected and regulated by highly flexible domains. Its structure, function and organization have been studied in several species, but the understanding of its overall assembly and dynamics remains challenging [23].
PDC is a large multienzyme system composed of three distinct catalytic components which work closely together (Table 1): E1, a heterotetramer of two α and two β subunits (E1α and E1β); E2, a homotrimer or heterotrimer of E2-E3BP, and E3, a homodimer. PDC also comprises a structural component: E3BP, formerly referred as protein X that mediates stable E3 integration into the E2 core of the complex [24]. Overall, the protein subunits of the PDC components are encoded by five genes, PDHA1, PDHB, DLAT, DLD, and PDHX. Moreover, important regulatory enzymes interact with PDC, which include E1-specific kinases (PDK) and E1 phosphatases (PDP) that regulate PDC activity by (de)phosphorylation [3–5, 25]. The global reaction catalyzed by PDC results in the irreversible decarboxylation of pyruvate, with the formation of reduced nicotinamide adenine dinucleotide (NADH), CO2 and acetyl-CoA. This series of coordinated reactions involve five coenzymes: thiamine pyrophosphate (TPP), lipoic acid (LA), CoA, FAD and NAD+.
Table 1.
Overview of the characteristics of human pyruvate dehydrogenase system (EC: 1.2.1.104)
| PDC Component: Enzyme (EC number) and Structural protein | Protein | Gene (human) | Oligomeric state/distribution in the multimeric PDC | Number of aminoacids (UniProtKB identifier) | Subcellular location | Protein function | Mutations/Diseases associated | Phenotype MIM (#) number |
|---|---|---|---|---|---|---|---|---|
| Pyruvate Dehydrogenase/Decarboxylase (EC:1.2.4.1) | E1α | PDHA1 | Heterotetramer (E1α2β2)/ periphery | 390 (P08559) | Mitochondrial matrix Nucleus |
Oxidative decarboxylation of pyruvate |
Pyruvate dehydrogenase E1α deficiency | 312170 |
| E1β | PDHB | 359 (P11177) | Pyruvate dehydrogenase E1β deficiency | 614111 | ||||
| Dihydrolipoamide Transacetylase/Acetyltransferase (EC:2.3.1.12) | E2 | DLAT | Homotrimer or heterotrimer/ inner core | 647 (P10515) | Mitochondrial matrix Nucleus | Acetylation of coenzyme A | Pyruvate dehydrogenase E2 deficiency | 245348 |
| Dihydrolipoamide dehydrogenase (EC:1.8.1.4) | E3 or DLD | DLD | Homodimer/ periphery | 509 (P09622) | Mitochondrial matrix Nucleus | Regeneration of oxidized lipoamide | Dihydrolipoamide dehydrogenase deficiency | 246900 |
| E3-binding protein | E3BP | PDHX | Undefined/ inner core | 501 (O00330) | Mitochondrial matrix Nucleus | Stable E3 integration into the E2 core of the complex | Pyruvate dehydrogenase E3-binding protein deficiency | 245349 |
In brief, activities of E1 and E2 generate acetyl-CoA, whereas the FAD/NAD+-dependent E3 mediates redox recycling [3, 26]. The first reaction is the irreversible decarboxylation of pyruvate catalyzed by E1 that requires the cofactor TPP. The second reaction is catalyzed by E2 and requires CoA to generate an acyl-CoA. The transfer of electrons from the dihydrolipoyl moieties of E2 to FAD and then to NAD+ is carried out by E3, a multifunctional oxidoreductase [27, 28]. Thus, the third reaction is catalyzed by this enzyme (DLD), that is responsible for the regeneration of lipoamide, reduction of NAD+, and oxidation of FADH2 [3] (Table 1). The properties and characteristics of the latter enzyme will be discussed in more detail throughout this work.
In eukaryotes, PDC comprises multiple copies of the three catalytic enzymes namely ca. 30 subunits of E1, 60 subunits of E2, and 12 subunits of E3. This proportion does not seem to be altered in case of upregulation of DLD [29] and the gene knockdown of E1α or E1β appears not to alter expression of other proteins [30, 31]. The entire complex (≈ 9.5 MDa) exhibits icosahedral symmetry [3]. Hence, the multienzyme assembly of human PDC represents a highly intricate process that will influence its function, structure, and regulation.
PDC activity is highly regulated, essentially through two different mechanisms: (1) the end-products, acetyl-CoA and NADH, may directly inhibit PDC, apparently by binding to the E2 and E3 active sites, respectively [26], or (2) via reversible phosphorylation of the E1 component [4, 5]. This modification is mediated by the regulatory enzymes: pyruvate dehydrogenase kinases (PDK1 to 4) and pyruvate dehydrogenase phosphatases (PDP1 and PDP2) [32]. The kinases (PDKs) phosphorylate the E1α subunit and inactivate PDC. The phosphatases (PDPs) revert the phosphorylated PDC-E1α to its active, dephosphorylated form [33]. Both the PDC kinases and the PDC phosphatases are Mg2+-dependent and respective mediated reactions are conditioned by ATP and/or ADP levels [5, 32, 33].
The activity of PDC is tightly influenced by the cellular levels of PDKs and PDPs in tissues, which are variable depending on normoxia or hypoxia conditions [34, 35] or different nutritional and disease states [3, 36–38]. Thus, PDKs and PDPs present exclusive tissue expression patterns, kinetic properties, and sensitivities to regulatory molecules. Dichloroacetate (DCA), omega-3 fatty acids and thiamine inhibit PDKs, thereby increasing PDC activity [26]. An upregulation of PDKs has been observed in patient´s tissues with metabolic diseases, suggesting that inhibition of these kinases might have beneficial effects in the treatment of metabolic diseases [34]. The influence exerted by PDPs and respective expression pattern have received much less attention compared to those of PDKs. PDP1, a Ca2+-sensitive isoform, is expressed in rat heart, brain, and testis whereas PDP2, which is a Ca2+-insensitive isoform, is abundantly expressed in rat kidney, liver, heart, and brain [34]. In the absence of Mg2+ or Ca2+ ions, the PDP is inactivated [39].
In addition, several endogenous molecules can also modify PDC activity, such as pyruvate, NAD+ and CoA, which lead to substrate activation of PDC by inhibiting PDKs [38]. The PDKs regulate PDC activity by responding to diverse allosteric modulators. High ratios of acetyl-CoA to CoASH and NADH to NAD+ represent signals of saturation of mitochondrial demand. Elevations of these ratios are strong activators of PDKs, shutting down PDC activity [26].
Besides phosphorylation, other post-translational modifications (PTMs) of PDC such as acetylation, glutathionylation, succinylation, or glycosylation have been referred [38] but the respective physiological implications need further elucidation.
The importance of PDC in disease pathophysiology is undisputed [26, 27], gaining an emergent attention in cancer biology [30, 37–41]. Clinical cases of hereditary PDC deficiency have been largely associated with mutations in the X-linked PDHA1 gene, encoding the E1α subunit, which reduce the catalytic activity of the enzyme or the stability of the overall complex. The phenotype typically presents between birth and infancy [6]. However, genetic deficiencies of PDC can also be caused by defects in the E1β, E2, E3 or E3BP subunits. [42].
Overall, a complex regulatory machinery synchronizes the expression, assembly, regulation and concerted function of all components of PDC. In this work, further focus will be given to E3 component because of its unique characteristics and relevance.
Dihydrolipoamide dehydrogenase (DLD) as a therapeutic target
In humans, the protein encoded by the DLD gene exists as a dimeric component (hE3) of PDC, and at least of four additional multienzyme systems. It was reported that dihydrolipoamide dehydrogenase (EC 1.8.1.4) (also abbreviated as DLD; LAD; LipDH; DLDD; DLDH; GCSL; PHE3; OGDC-E3), exists as a free enzyme in vivo and it is the most abundant flavoprotein in muscle and brain mitochondria [43]. DLD represents a resourceful enzyme with multiple roles in mitochondrial metabolism and redox equilibrium. It has been described as a moonlighting protein based on its ability to perform mechanistically distinct functions (oxidoreductase, diaphorase, protease, DNA complex formation) [43–45]. It is characterized by a ubiquitous expression in heart, kidney, liver, brain, and many other tissues. This protein has been also identified in human and rat serum, where no 2-oxoacid dehydrogenase complexes are detected, but the biochemical properties of serum E3 remain to be truly described [28].
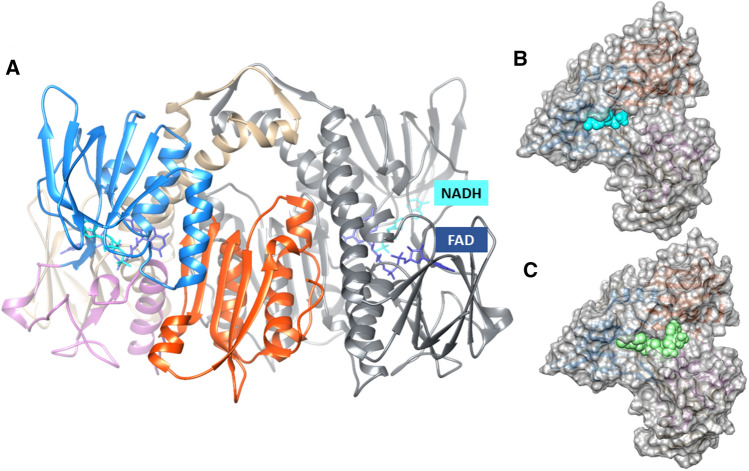
The crystal structures for hE3 have been published, including those for specific disease-causing mutations [46–53]. Structurally, the mature form of hE3 subunit (474 amino acid residues; 54 kDa) presents four domains: the N-terminal FAD-binding domain (1–149), the NAD+/NADH-binding domain (150–282), the central domain (283–350), and the C-terminal interface domain (351–474) [48–51] (Fig. 1). DLD is a protein homodimer that catalyzes the oxidation of dihydrolipoamide, using one FAD prosthetic group bound very tightly but non-covalently to each monomer of E3 (subunit), and a transiently bound substrate, NAD+ [43, 48]. Nevertheless, the respective oligomeric state can change from dimeric to monomeric or tetrameric depending on the pH in the mitochondrial matrix [44, 54]. Thus, modifications of pH may alter oligomerization and DLD catalytic properties with potential consequences to the structure and function of the energy-producing mitochondrial complexes that contain this enzyme.
Fig. 1.
Structural representation of human dihydrolipoamide dehydrogenase (hE3) homodimer (A). The position of NADH (in cyan) into the structure of a hE3 monomer is shown (B). Best docking poses of valproyl-CoA (in green) into a hE3 monomer (C). A One monomer of hE3 is coloured according to the protein domains: the FAD and NAD binding domains in tan and blue, respectively, the central domain in purple and the interface domain in red; the second hE3 monomer is coloured in grey; the FAD cofactor and the NADH are depicted in van der Waals representation and in dark blue and cyan, respectively. Figure was created with UCSF Chimera and by superposing the PDB structures 1ZMC and 3RNM. The models (B, C) were built with Autodock Vina and using the human E3 PDB ID 6I4Q.
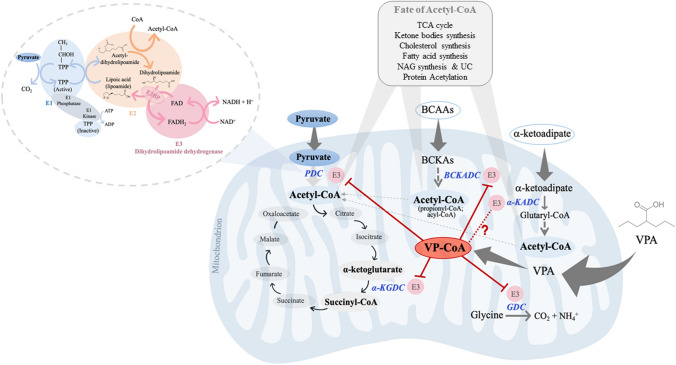
Beyond integrating PDC, E3 (or DLD) also participates in the mitochondrial α-ketoacid dehydrogenases complexes α-KGDC, BCKADC, and GDC commonly designated as GCS [55]. More recently, it was reported that hE3 is also part of the α-KADC, responsible for the oxidative decarboxylation of α-ketoadipate to glutaryl-CoA, a product of the degradation pathway of l-lysine, l-hydroxylysine, and l-tryptophan [56, 57] (Fig. 2). All these enzyme systems are localized in the mitochondrial compartment and share similarities in structure and the same cofactor dependency, even though they participate in distinct metabolic pathways.
Fig. 2.
VPA interference with E3 (DLD) and E3-dependent multienzyme systems may have impact in mitochondrial metabolic pathways generating acetyl-CoA. The role of FAD/NAD+-dependent E3 on redox recycling is depicted on overall reactions of the mitochondrial oxidation of pyruvate to acetyl-CoA involving the PDC components, as an example among the α-ketoacid dehydrogenase family. The mitochondrial activities of PDC, α-KGDC, BCKADC, and GDC may be altered in the presence of VPA, namely through valproyl-CoA (VP-CoA)-induced effect on E3 proteins. The scheme highlights the role of PDC, interlinking cytosolic glycolysis (pyruvate) to the TCA cycle (acetyl-CoA). The question mark underlines the lack of evidence for this interaction. PDC pyruvate dehydrogenase complex; E1 Pyruvate dehydrogenase; E2 dihydrolipoamide transacetylase; E3 dihydrolipoamide dehydrogenase; E3BP E3-binding protein; TPP thiamine pyrophosphate; NADH/NAD+ nicotinamide adenine dinucleotide (reduced/oxidized forms); FADH2/FAD flavin adenine dinucleotide (reduced/oxidized forms); α-KGDC α-ketoglutarate dehydrogenase complex; BCAAs branched-chain amino acids; BCKAs branched-chain α-keto acids; BCKADC branched-chain α-keto acid dehydrogenase complex; GDC glycine decarboxylase complex; α-KADC α-ketoadipate dehydrogenase complex. TCA tricarboxylic acid; NAG N-acetylglutamate; UC urea cycle
In the corresponding multienzyme complexes, the common E3 is responsible for transferring two electrons from dihydrolipoamide to NAD+, mediated by FAD which is reduced to FADH2. As depicted in Fig. 2 for PDC, the E3 completes the final step in a series of synchronous co-operating catalytic reactions where it becomes a key enzyme to maintain the metabolic flow. The global reaction catalyzed by DLD (E3) is described as [58]:
E3 catalyzes the reduction of NAD+ to NADH, transforming dihydrolipoamide into lipoamide (forward direction). The catalytic reaction occurs through the redox center of the enzyme, which is constructed by the combination of a disulfide bridge coupled to the flavin ring of the cofactor FAD [59]. The reduced form FADH2 is produced transiently in the first half-reaction and oxidized back to FAD, reducing NAD+ to NADH in the last half-reaction (Fig. 2) [59]. Then, NADH can feed into the electron transport chain for OXPHOS and ATP synthesis.
E3 is present in the multienzyme complexes mentioned above as a homodimer, where it acts as an oxidoreductase. However, the enzyme can exist as a monomer and play a distinct role, namely acting as a protease [27]. It has been shown that in vitro, E3 can also act as a diaphorase, transferring electrons from NADH to electron acceptors, such as cytochrome c, ubiquinone, and nitric oxide [60].
It is relevant to note that E3 binding within the α-KGDC is approximately 30 times weaker than in the PDC, and the interaction with the remaining components of α-KGDC may be further weakened in acidosis [43]. These observations lead us to speculate that when a suboptimal quantity of hE3 is present, as in cases of E3-deficiency or metabolic acidosis, the function of PDC will be more resilient and less prone to be affected than α-KGDC.
The molecular pathomechanisms of hE3 deficiency were revised under the recently published crystal structures for hE3 [55, 61]. E3 deficiency is a rare autosomal recessive genetic disorder, with symptoms frequently arising in neonatal age (early-onset) and that may lead to death. Affected children who live beyond the first 2–3 years may develop growth deficiencies and neurological abnormalities. The highest carrier rate has been found among Ashkenazi Jewish population with a disease frequency of 1:35,000–1:48,000. This disorder manifests mostly by neurologic, cardiac, and (adult-onset) hepatic symptoms. The phenotype includes, among others, hyperammonemia, encephalopathy, seizure, liver dysfunction, lactate acidosis, hypoglycemia, Leigh syndrome, and developmental delay [62]. The variable phenotype and clinical characteristics of DLD deficiency may be due to the involvement of E3 in several complexes, as described above, and consequences of their affected activities. Interestingly, although DLD also participates on the GCS where it is known as the l-protein, pathogenic mutations in the DLD gene do not appear to impair the function of this system in vivo [62]. Nowadays, there is no cure for this disease due to the genetic condition, and management may be achieved through dietary restrictions avoiding fasting/catabolism and liver-toxic medications, correction of metabolic acidosis, and also by administration of flavins, LA and TPP [55].
In terms of pathophysiology, E3 dysfunction can lead to the accumulation of pyruvate, lactate or branched-chain amino acids (BCAAs) in plasma and rise of branched-chain α-ketoacids (BCKAs) in urine [62, 63]. In addition, some studies have explored the involvement of DLD on redox status. It has been described that DLD can either enhance or attenuate production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), depending on experimental or pathophysiological conditions [27]. Studies in vitro using isolated mitochondria have shown that 2-oxoacid dehydrogenases may produce superoxide/H2O2, reporting the maximum capacities as α-KGDC > PDC > BCKADC [64]. When E3 is fully functional it can scavenge nitric oxide, acting as a protective enzyme [65]. However, dysfunction of E3 may lead to an overproduction of ROS which, in turn, causes oxidative stress [66, 67]. The ROS-generating activity of the intact α-KGDC (and the PDC) had been attributed to the E3 component, potentiated by acidosis, but E1 and E2 have been also ascribed [68]. PDC seems to represent a much weaker source of ROS than α-KGDC, but PDC is also sensitive to ROS, and its functional integrity is important for neuronal longevity [68–70].
Oxidative stress has been associated with ageing and brain dysfunction. It is noteworthy that impairment of human α-KGDC activity in brain represents a common sign in several age-associated neurodegenerative diseases, including Alzheimer’s and Parkinson’s diseases, as well as Friedreich’s ataxia [69]. Specific antioxidants, flavins or lipoic acid are proposed for therapy [43]. However, studies in rodents suggest that senescence is not associated with clear changes in the activity of DLD or its susceptibility to inactivation by mitochondrial oxidative stress [71]. Interestingly, oxidative stress is a pathological mechanism of type 2 diabetes and it was proposed that chronic E3 inhibition could serve as a protective approach to manage oxidative stress in this specific disease [27]. A recent study in cancer cells suggests that DLD is closely linked to ferroptosis, induced by cystine deprivation or import inhibition [72], where the α-KGDC-dependent oxidative stress could profoundly affect the redox state of cancer cells [72]. Treatments with bioengineered DLD in a subcutaneous melanoma mice model resulted in significant tumor inhibition [73].
The understanding and the targeting of mitochondrial pyruvate metabolism for cancer therapies remains a challenge [74]. Considering the above-mentioned studies, whether E3 is a potentially “druggable” target for any disease is one hypothesis that has not been fully explored yet.
Valproic acid as a potential inhibitor of DLD and DLD-dependent complexes
Studies on design and development of potential specific E3 modulators have been scarce. There are several known activators: TPP, pyruvate, DCA, and LA, which are recognized inhibitors of PDKs that thus activate PDC and in turn the E3 component [26, 75], beyond the known E3 cofactors, NAD+ and FAD. However, few molecules were reported as specific inhibitors of E3 activity. Besides the inactivating effect of some inorganic species such as zinc [76, 77], arsenite [78] or phosphine gas [79], the inhibitory effect of two small organic molecules were described, the 5-methoxyindole-2-carboxylic acid (MICA) [80] and valproic acid (VPA) metabolites [81].
The first inhibitor, MICA, has been described as a hypoglycemic agent with potential antidiabetic properties [80]. Our focus will be on the second, VPA (2-n-propyl-pentanoic acid), also used as a valproate salt of sodium or magnesium. It is a well-known drug in clinical practice for decades, and one of the most prescribed antiepileptic drugs in the world.
VPA was synthesized as a derivative of valeric acid, a component of the herbal medicine Valeriana officinalis [82]. It is a C8-branched medium-chain fatty acid with significant pharmacological effects, such as antiepileptic, anticonvulsant and mood-stabilizing drug [83, 84]. Its anticonvulsant effect may be related to an increase in gamma-aminobutyric acid (GABA) levels in brain and subsequent blockage of VDAC [85]. Valproate is also included in various clinical guidelines as one of the recommended therapies for migraine prophylaxis [86, 87]. In addition, it has integrated various clinical trials for cancer therapy.
Herein, some evidence that intersect valproic acid, as a potential inhibitor of E3, with pyruvate metabolism, PDC function or other E3-dependent complexes, will be described.
VPA teratogenicity and hepatotoxicity
VPA is a first-line option in the treatment of generalized epilepsies, unless safety-related contraindications are present [82]. There are two major potential problems that still remain, teratogenicity and hepatotoxicity [88–91], but other VPA adverse drug reactions have been systematically reviewed [92]. The mechanisms by which the drug exerts these unwanted effects are still unclear but it is consensual that interactions with mitochondrial energy metabolism and/or inhibition of histones deacetylases (HDAC) may play an important role. Exposure to VPA in utero has been reported to cause severe birth defects and impaired postnatal development. Currently, available evidence suggests that the risk of congenital malformation after VPA exposure is around 11% up to 24% depending on the dose [91]. Moreover, VPA induces mild to severe hepatic dysfunction characterized by microvesicular steatosis, weight gain, insulin resistance, and hyperammonemia [92]. Hepatotoxicity is a serious and complicated side effect that can become lethal, especially in children. Some risk factors for severe VPA-induced hepatotoxicity are young age, developmental delay, congenital metabolic disorders (namely mitochondrial enzyme deficiencies) or even ketogenic diet [93–95]. Numerous studies reported over the last 5 decades, the occurrence of a Reye-like syndrome in epileptic patients treated with valproic acid (VPA), an adverse event at hepatic level due to mitochondrial dysfunction [94].
Several molecules have been investigated in coadministration with VPA with the aim to minimize adverse effects of the drug. Carnitine is one of the most important examples, showing protective effects in animal models [95–97]. Moreover, drug-induced hepatotoxicity has been shown to be accompanied by an inflammatory reaction and ellagic acid, a polyphenol, has revealed antioxidant, antiviral, and antibacterial properties. It was claimed that this acid, when administered orally, protects liver from VPA toxicity [98]. The same was observed for melatonin, which also acts as an antioxidant, and protective agent against hepatotoxicity caused by VPA [99].
VPA metabolism and mitochondrial dysfunction
The detailed mechanisms that explain why VPA causes teratogenicity and hepatotoxicity have not yet been fully established. It is known that this drug induces pleiotropic effects and interferes with several pivotal metabolic pathways namely pyruvate-driven oxidative phosphorylation, FAO and urea synthesis [10, 11]. In this context, it is relevant to understand the potential implications on PDC, which interconnects these pathways, on acetyl-CoA levels and redox balance.
The interactions between VPA metabolism and bioenergetics were clarified with the identification of the complete β-oxidation of the drug in mitochondrion [100]. Before this report, the reactions mediated by microsomal-linked enzymes attracted most studies on VPA metabolism. Then, based on its unique properties of branched-chain fatty acid, reports on the extra-mitochondrial formation or mitochondrial accumulation of valproyl-CoA (VP-CoA) [101, 102] and the interaction with BCAA oxidative pathway [103, 104], gave unequivocal evidence that this drug has important effects on mitochondrial metabolism.
Studies in vitro showed that VPA can clearly interferes with pyruvate transport, using mitochondrial membranes [105] or brain mitochondria [106], and with pyruvate oxidation rate [11]. The clear inhibitory effects of VPA on pyruvate-, α-ketoglutarate-, and glutamate-driven oxygen consumption and ATP synthesis were explained through the hypothesis of a DLD-targeted inhibition [81, 107]. In fact, the major mitochondrial derivative of VPA, valproyl-CoA, was shown to induce a mild inhibition of DLD activity, using purified α-KGDC and PDC as enzyme sources [81]. Valproyl-CoA was shown to be the most abundant CoA ester of the drug, formed in mitochondrial matrix, with higher resistance to hydrolysis than endogenous acyl-CoAs of linear-chain fatty acids [101, 102]. This fact suggests the longer residence time in mitochondria of valproyl-CoA, with potential sequestration of free CoA, although it was shown that valproyl-CoA is also a substrate of short branched-chain acyl-CoA dehydrogenase (SBCAD) [103, 104]. Figure 1C depicts the best molecular docking of valproyl-CoA on DLD, suggesting a localization of this ligand on the NAD+ binding site of the protein. Thus, it is legitimate to hypothesize that a potential targeted effect on E3 may occur. Interestingly, in rat brain mitochondria, the pyruvate and 2-oxoglutarate oxidation rates appear to be unaffected by VPA [107]. This discrepancy is consistent with the report that valproyl-CoA could not be detected in rat brain, using animals exposed to the drug, but further studies in brain are needed [108].
VPA metabolism, PDC, and other E3-dependent complexes
In human, the putative interaction between valproyl-CoA and DLD activity, remains unclear. This metabolite is strictly intracellular and ethical considerations are involved in the sample collection and challenging study of drug metabolites levels in patient tissues. Evidence of this interaction is provided when VPA administration triggers specific metabolic abnormalities. As a paradigm, there is a case report of a child under neurodevelopment therapy sessions, with uncontrolled seizures despite antiepileptic medication [109]. The observed increase in BCAAs, ornithine, blood lactate, pyruvate, and ammonia were suggestive of an inherited metabolic disorder, the E3-dihydrolipoamide dehydrogenase deficiency. Metabolomic approaches of serum in epileptic patients treated with VPA sustain this pattern of metabolites changes which may reflect disturbances on respective metabolic pathways [110, 111].
VPA-treated patients have been reported to show higher levels of glycine in serum and especially in urine, suggesting a clear reduced GCS activity [110, 112]. The hyperglycinemia and hyperglycinuria, associated with VPA treatment, are most likely due to enzymatic inhibition of GCS [112, 113]. Since E3 is a component of the GCS structure (Fig. 2), it can be hypothesized that drug-induced changes on glycine levels may be assigned to E3 inhibition within GCS. As far as we know none of these reports linked the inhibitory effects of VPA on GCS with a potential decrease in E3 activity or protein levels induced by valproyl-CoA [81]. Still, glycine levels are normal in almost all patients with E3 deficiency, with the exception of one report of reduced GCS in a liver sample of a E3 deficient patient [114, 115].
Patients harboring mutations in the DLD gene, encoding the E3 proteins of all α-ketoacid dehydrogenase systems, presented reduced activity of the α-KGDC and BCKADC complexes [62]. Glycine accumulation has not been reported in individuals with PDC or α-KGDC deficiency [116, 117]. Overall, further studies will be essential to unveil the mechanisms underlying VPA-associated hyperglycinemia and inherent dysfunction of E3 or GCS.
BCAAs, such as leucine, isoleucine, and valine, are essential amino acids that are not only necessary for protein synthesis but also play important physiological roles. Their catabolism is controlled by the mitochondrial BCKADC [118, 119]. The administration of VPA in vivo clearly interacts with BCAAs catabolism, where first reactions correspond to a cycle of β-oxidation, explained also by their similar branched carbon-chain structures [81, 103, 104]. It has already been demonstrated that patients receiving VPA showed an increase in serum levels of BCAAs and their intermediates as well as an increase in urine levels of specific BCAAs derivatives [104, 120, 121]. The purified mammalian BCKADC is essentially devoid of the E3 component, so the absence of E3 is associated with the low affinity of the subunit-binding domain of human BCKADC for hE3 [51]. Thus far, there is no X-ray crystal structure for any of the 2-oxo acid dehydrogenase complexes in their intact state, possibly due to their size and flexible attachment of the E1 and E3 components to the core [122, 123]. The hypothesis of VPA interference with BCKADC is reinforced by analogy with the reported inactivation of purified mammalian α-KGDC by valproate or its metabolites [124]. Therefore, it can be speculated whether there is an altered activity of both complexes, namely through valproyl-CoA mediated effect on E3 (Fig. 2) but further studies are needed because little is known about this interaction.
The role of α-KGDC in human brain is very important to neurotransmission and it is fundamental as a regulatory enzyme within the tricarboxylic acid cycle (TCA). A competitive inhibition of purified beef brain α-KGDC by valproyl-CoA, was demonstrated in vitro [124]. In silico studies supported the prevalent hypothesis of a minor involvement of α-KGDC in the anticonvulsant activity of VPA [125]. DLD was shown to be present in neurons and glia as well as non-neuronal cells in various regions of human brain [126]. α-KGDC is sensitive to ROS and inhibition of this enzyme may be critical in metabolic deficiencies induced by oxidative stress [127]. A decline in α-KGDC activity is associated with various neurodegenerative diseases but the molecular mechanism(s) linking this multienzyme complex to neurodegeneration are far from being elucidated [126, 127].
The subcellular communication between cytosolic glycolysis and mitochondrial FAO is ensured through PDC activity and by common metabolites such as acetyl-CoA and the NAD+/NADH ratio. The intracellular NAD+/NADH ratio contributes to the activation/inhibition of PDC by affecting DLD activity, which is critically dependent on pH [54, 128]. Interestingly, administration of VPA in vivo has been reported to interfere with liver NAD+ concentration affecting NAD+ availability or their precursors at several levels [110, 129]. These alterations show a potential disruption of NAD+ cofactor homeostasis, but the transient VPA-associated NAD+ depletion may involve distinct unknown mechanisms [110]. Therefore, a significant decrease of matrix NAD+ pool has an inhibitory effect on enzymatic reactions that require NAD+ as a cofactor and, as a consequence, the activity of PDC may be compromised [130]. A recent study in E. coli has shown that PDC sensitivity to NADH resides in the E3 component, as only this enzyme interacts with NAD+ as a substrate [131]. The ability of the E3 component to regenerate oxidized lipoic acid is controlled by the NAD+/NADH ratio within the mitochondria. When the availability of NAD+ is diminished, FADH2 within the E3 component can readily be oxidized by O2 generating a semiquinone (FADH•) and superoxide (O2•−) [132]. The emergent role of lipoic acid metabolism has been recently reviewed, underlining the importance of lipoylation-dependent enzymes to mitochondrial redox regulation [132]. Furthermore, the role of SIRT4 (Sirtuin 4) as a cellular NAD+-dependent lipoamidase was shown to regulate PDC and this catalytic efficiency for lipoyl-lysine modifications seems to be superior to its deacetylation activity [133].
The activity of α-KADC on lysine degradation also requires lipoic acid and depends on E3 role. Recently, a functional and regulatory crosstalk between the human α-KGDC, the regulatory enzyme at the TCA cycle mentioned above, and this novel α-KADC system (participating on the catabolism of l-lysine, l-hydroxylysine and l-tryptophan) has been reported [134]. Remarkably, the “backbone” of lipoic acid is octanoic acid, the linear-chain eight-carbon fatty acid [117]. Whether and how VPA, the branched-chain eight-carbon fatty acid, interferes with this lipoic acid- and DLD-dependent enzyme remains an unanswered question that, in our perspective, deserves elucidation (highlighted in Fig. 2).
In summary, VPA and/or its intermediates as valproyl-CoA, affect mitochondrial energy metabolism (Fig. 2) through multiple pathways or targets where interference with PDC activity, more specifically, with its E3 component, may play a pivotal role in mechanisms of drug-induced liver toxicity or in other prenatal adverse effects.
Nuclear acetyl-CoA and valproate interactions as an inhibitor of KDAC
Nuclear acetyl-CoA and PDC
The identification of PDC in the nucleus links acetyl-CoA formation with lysine acetyltransferases/deacetylases (KATs/KDACs) activities, where histones are the major protein target. Therefore, we will discuss VPA-induced alterations on histone acetylation status, to bring some light into the yet obscure mechanisms linking metabolism and epigenetics. Further research is needed to clarify whether VPA-induced effects on E3 component of mitochondrial PDC, may also occur at the level of nuclear PDC, creating an additional control mechanism of substrate availability for histone acetylation enzymes.
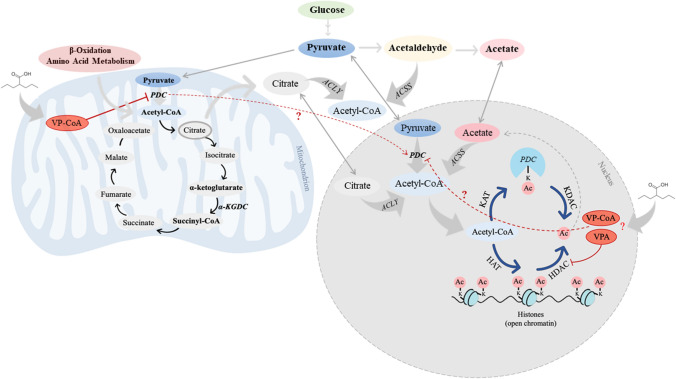
The reversible acetylation of histones is one of the main modifications with crucial importance for epigenetic control of gene expression [135]. Lysine acetylation is a reversible and conserved PTM that changes the charge on lysine residues and thereby modifies protein structure. PTMs may occur not only in nuclear histones but also in numerous non-histonic proteins such as enzymes of intermediary metabolism present in different cellular organelles including mitochondria. Concerning histonic proteins, acetyl-CoA is bound to histone tails by histone acetyltransferase (HAT) altering chromatin dynamics and thus gene expression [136]. Thereby, acetylation is directly dependent of acetyl-CoA levels, further indicating that acetyl-CoA is a limiting factor for several acetylation events and a crucial regulatory metabolite [136, 137]. And because organelle membranes are impermeable to this large molecule, the mitochondrial and non-mitochondrial pools of acetyl-CoA are independently produced. Mitochondrial acetyl-CoA is generated by PDC, as well as by FAO and amino acid metabolism (Fig. 3). On the other hand, the non-mitochondrial pool of acetyl-CoA is produced in the cytosol by ATP citrate lyase (ACLY) and acetyl-CoA synthetase (ACSS), and in the nucleus by the nuclear PDC (Fig. 3) [138, 139]. Thus, acetyl-CoA (Figs. 2 and 3) has a dual role as the acetyl-donor for protein acetylation (histones and non-histones) and as a central regulatory metabolite connecting acetylation, metabolism and epigenetics [140, 141].
Fig. 3.
Acetyl-CoA production and dynamic acetylation and deacetylation of histones. Acetyl-CoA is produced in the cytosol and nucleus by ATP citrate lyase (ACLY) using citrate exported from TCA cycle and by acetyl-CoA synthetase (ACSS) from acetate. Mitochondrial PDC can translocate to the nucleus and produce nuclear acetyl-CoA directly from pyruvate. Acetyl-CoA provides the necessary acetyl groups to enable the protein acetylation reactions. HAT histone acetyltransferase; KAT lysine acetyltransferase; HDAC histone deacetylase; KDAC lysine deacetylase; Ac acetate; CoA Coenzyme A; VP-CoA valproyl-CoA; VPA valproic acid
It was shown that all protein components of the mitochondrial PDC are also present and functional in the nucleus, since the knockdown of nuclear PDC in isolated functional nuclei decreased the de novo synthesis of acetyl-CoA and acetylation of core histones [8, 35, 142]. Though the dynamic translocation of mitochondrial PDC in the nucleus is yet to be fully conceived [143], its correlation with acetylation of histones underscores its role in epigenetic regulation [8, 35, 143]. The inhibition of nuclear PDC leads to hypoacetylation of core histones [144]. PDC has been found to enter the nucleus during conditions of mitochondrial stress, in response to growth factors, mitochondrial inhibitors (e.g. rotenone) or oncogenic signaling [8, 35, 140]. This suggests a potential role in disease states that involve proliferative signals or mitochondrial dysfunction. Interestingly, inhibition of ACLY has been shown to suppress tumorigenesis [145, 146]. Whether mitochondrial or cytosolic valproyl-CoA is translocated in the nucleus and interferes with nuclear DLD and PDC are hypotheses under investigation (Fig. 3).
Nuclear KDAC and VPA
In general terms, acetylation of proteins is controlled by the following two groups of enzymes (Fig. 3): KATs and KDACs (including HDACs) [137, 138, 147]. KATs may promote the transference of an acetyl group from acetyl-CoA to the ε-amino group in the side chain of lysine residues. The reverse reaction, the deacetylation process, is accomplished by KDACs which use Zn2+ as a cofactor, or by sirtuins which are a specific group of NAD+-dependent KDACs also present in cytosol and mitochondria [143, 148]. Considering the pivotal role of KDACs on the regulation of histone acetylation and gene expression, compounds capable of inhibiting the activity of these enzymes may have therapeutic potential [147, 149].
VPA was identified as an inhibitor of class I and II HDACs [83, 150] Since then, VPA has been recognized as a KDAC inhibitor (KDACi) (Fig. 3), which is related to its potential anti-cancer and neuroprotective effects [151, 152]. These properties have opened new perspectives for the repurposing of this drug, especially in cancer therapy, with inclusion in numerous clinical trials and emerging studies [152–154]. Whether valproyl-CoA exists in the nucleus and interferes with HDAC activity are unanswered questions and hypotheses to elucidate (Fig. 3).
Acting as an epigenetic drug, VPA increases the expression of many genes involved in various cancer-related processes, leading to differentiation, inhibition of cell growth and increased apoptosis and immunogenicity [151, 154, 155]. The drug leads to a decrease in metastatic and angiogenic potency of tumor cells [152]. It also increases the acetylation of histone proteins and relaxes chromatin conformation [156]. VPA down-regulates HDAC activity in teratocarcinoma and neuroblastoma cells [152]. Studies have shown that VPA inhibits HDAC activity in human hepatocellular carcinoma (HCC) cells [153], as well as selectively inhibits HCC cells proliferation without harming normal hepatocytes [155]. VPA sensitizes cancer cells against radiation, thus making it a beneficial radio-sensitizer in breast cancer radiotherapy [157]. This drug is being tested in phase II clinical trial for the treatment of leukemia and solid tumors [158]. Curiously, KDAC inhibition by systemic administration of VPA has been shown to protect neurons by modulating neuroinflammation and to improve Parkinson’s disease-like behaviors [159].
Metabolic alterations on acetyl-CoA levels have also been shown to be associated with metastatic phenotype in several cancers [160]. Additionally, high nuclear acetylation levels have been observed in cancer cells resulting from the increased activity of acetyltransferases and the ectopic synthesis of acetyl-CoA in the nucleus [161]. For metabolic reprogramming in cancer metabolism, PDC is highly regulated through transcription factors and oncogenes in most cancers [30, 31]. PDC acetylation has been reported to inactivate the protein complex, leading cancer cells to depend on glycolysis for cell survival [162, 163]. Inhibition of PDC activity has been reported to contribute to the malignant phenotype in human head and neck squamous cell carcinoma [164]. Acetylation inhibits PDC through increased expression of PDK-1, which results in inhibitory phosphorylation of the PDC-E1α subunit [164, 165]. On the other hand, PDC-E1α and the PDC activator, PDP1, are often overexpressed at both transcript and protein levels in prostate tumors [166].
While the involvement of PDC-E1α subunit in cancer metabolism is often studied, the role of DLD is seldom investigated with PDC activity in metabolic reprogramming in cancer [72, 73, 167] and to our knowledge, no intersections exist with VPA-associated epigenetic interactions. To conclude, regulation of PDC in different organs and tissues is crucial in proliferative cancers, but it must be emphasized that it is also prominent in age-related diseases such as obesity, diabetes, neurological disorders including Alzheimer’s and Parkinson’s diseases [136].
Mitochondrial metabolism, epigenetics, and VPA
The importance of protein (and histones in particular) acetylation was highlighted in previous paragraphs. Nevertheless, it is relevant to underline that the modifications of lysine residues in proteins are not limited to acetylation, since ubiquitination, methylation, glutarylation, succinylation or other acyl groups deserve great attention. Much fewer selective modulators for these modifications are known as compared with acetylation [147]. Importantly, a tight link between metabolism and epigenetic regulation of gene expression by succinylation and glutarylation was recently reviewed [134], which is clearly related with DLD-dependent complexes. The signaling role of mitochondrial metabolites to control cell fate and function is well-recognized and how the metabolites variations in their abundance may control physiology and disease were recently reviewed [141].
Therefore, the hypothesis of VPA-induced effects on all E3-dependent multienzyme complexes may well interfere with the levels of relevant metabolites such as 2-ketoglutaric, glutaric and succinic acids with impact at epigenetic level. Novel findings concerning these hypotheses will certainly be reported in a near future.
Conclusions and perspectives
The underlying genetic background of a patient and the clinical condition that justifies VPA administration, do not facilitate the clear separation of drug effects per se (pharmacological or toxic) from cellular metabolism in vivo. The hepatic phenotypic presentation of certain cases of DLD gene mutations associated with Reye-like syndrome [168, 169] has clear similarities with reported cases of VPA-associated hepatotoxicity [92–94]. In these specific clinical cases of E3 deficiency confirmed by genome analysis, biochemical investigations were normal and only histologic examination of liver and muscle showed mild lipid inclusions [168, 169]. Authors conclude that DLD deficiency should be considered in patients with Reye-like syndrome or liver failure even in the absence of suggestive biochemical findings. These aspects underscore the potential liver susceptibility of VPA-induced inhibitory effect on DLD function and reinforce the great importance of considering an inborn error of metabolism as a genetic underlying cause for the VPA-associated hepatic toxicity [10].
Epilepsia is emerging as a metabolic disease, where mitochondrial dysfunction or energy metabolism dysfunction is associated with epileptic seizures [170, 171]. Therefore, an in-depth understanding of metabolism and its role in pathophysiology is essential not only to advance on novel therapeutic targets, but also to improve the safety of existing drugs.
In conclusion, the mechanisms of VPA-associated inhibition of pyruvate oxidation and E3-dependent multienzyme systems are multiple and elusive. The implications of a dual inhibition on PDC and FAO on limiting mitochondrial acetyl-CoA production were discussed. Whether the VPA-associated interaction with the E3 component of PDC or α-KGDC can be extrapolated to the potential inhibition of BCKACD, GDC and 2KADC complexes remains to be elucidated. The characteristic hyperglycinuria of VPA-treated patients supports the hypothesis of an E3-decreased activity on GDC. The relevance of VPA is undisputed, with a growing spectrum of pharmacologic claims in neuropsychiatric or other disorders, including as coadjuvant in anti-cancer combinatory schemes. However, the potential hepatotoxicity or teratogenicity needs to be further addressed, where the role of nuclear E3/PDC and drug interaction as a recognized KDAC inhibitor are not fully elucidated. Considering the published results and valuable reviews, now revisited, it is clearly demonstrated that VPA stands as a powerful metabolic and epigenetic modulator through mitochondrial function. The consequences to LA metabolism and whether and how VPA alters the pattern of protein acetylation or protein lipoylation are still unclear. Additional studies are required to clarify the importance of E3 as a potential drug target [172] and of the molecular bases of iatrogenic effects of VPA, with added interest to other KDACi or other FAO inhibitors. This future perspective will provide novel insights into the regulatory mechanisms of acetyl-CoA dependent mitochondrial energy homeostasis involving protein PTMs, enzyme activity, and gene expression.
Acknowledgements
Financial support was provided by Fundação para a Ciência e a Tecnologia (FCT, Portugal), through iMED.ULisboa: UID/DTP/04138/2019-2020.
Abbreviations
- Acetyl-CoA
Acetyl-Coenzyme A
- ACLY
ATP citrate lyase
- ACSS
Acetyl-CoA synthetase
- ALT
Alanine aminotransferase
- ATP
Adenosine triphosphate
- BCAA
Branched-chain amino acid
- BCKA
Branched-chain α-ketoacid
- BCKADC
Branched-chain α-ketoacid dehydrogenase complex
- DCA
Dichloroacetate
- E1
Pyruvate dehydrogenase (component E1)
- E2
Dihydrolipoamide transacetylase (component E2)
- E3/DLD
Dihydrolipoamide dehydrogenase (component E3)
- E3BP
E3-binding protein
- E3BD
E3-binding protein domains
- FAO
Fatty acid β-oxidation
- GCS
Glycine cleavage system
- GDC
Glycine decarboxylase complex
- HAT
Histone acetyltransferase
- HCC
Hepatocellular carcinoma
- HDACi
Histone deacetylase inhibitor
- hE3
Human E3
- KAT
Protein lysine acetyltransferase
- KDAC
Protein lysine deacetylase
- KDACi
Protein lysine deacetylase inhibitor
- α-KADC
α-Ketoadipate dehydrogenase complex
- α-KGDC
α-Ketoglutarate dehydrogenase complex
- LA
Lipoic acid
- LDH
Lactate dehydrogenase
- MICA
5-Methoxyindole-2-carboxylic acid
- MPC
Mitochondrial pyruvate carrier
- NAD(H)
Nicotinamide adenine dinucleotide (reduced form)
- NAG
N-Acetylglutamate
- OAA
Oxaloacetate
- OXPHOS
Oxidative phosphorylation
- PC
Pyruvate carboxylase
- PDC
Pyruvate dehydrogenase complex
- PDK
Pyruvate dehydrogenase kinase
- PDP
Pyruvate dehydrogenase phosphatase
- PTM
Post-translational modification
- TCA
Tricarboxylic acid
- TPP
Thiamine pyrophosphate
- UC
Urea cycle
- Valproyl-CoA
Valproyl-Coenzyme A
- VDAC
Voltage-dependent anion channel
- VPA
Valproic acid
Author contributions
IFD and MFBS wrote the manuscript and performed the literature search. All authors critically revised the versions of the manuscript. All authors read and approved the final manuscript.
Funding
Financial support was provided by Fundação para a Ciência e a Tecnologia (FCT, Portugal), through iMED.ULisboa: UID/DTP/04138/2019-2020.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
All authors declare no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Melser S, Lavie J, Bénard G. Mitochondrial degradation and energy metabolism. Biochim Biophys Acta. 2015;1853:2812–2821. doi: 10.1016/j.bbamcr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun. 2017;482:426–431. doi: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 3.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behal RH, Buxton DB, Robertson JG, Olson MS. Regulation of the pyruvate dehydrogenase multienzyme complex. Annu Rev Nutr. 1993;13:497–520. doi: 10.1146/annurev.nu.13.070193.002433. [DOI] [PubMed] [Google Scholar]
- 5.Harris RA, Bowker-Kinley MM, Huang B, Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Advan Enzyme Regul. 2002;42:249–259. doi: 10.1016/s0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 6.Sperl W, Fleuren L, Freisinger P, Haack TB, Ribes A, Feichtinger RG, et al. The spectrum of pyruvate oxidation defects in the diagnosis of mitochondrial disorders. J Inherit Metab Dis. 2015;38:391–403. doi: 10.1007/s10545-014-9787-3. [DOI] [PubMed] [Google Scholar]
- 7.De Boer VCJ, Houten SM. A mitochondrial expatriate: nuclear pyruvate dehydrogenase. Cell. 2014;158:9–10. doi: 10.1016/j.cell.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of Acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 9.Dejligbjerg M, Grauslund M, Litman T, Collins L, Qian X, Jeffers M, et al. Differential effects of class I isoform histone deacetylase depletion and enzymatic inhibition by belinostat or valproic acid in HeLa cells. Mol Cancer. 2008;7:70. doi: 10.1186/1476-4598-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva MFB, Aires CCP, Luís PBM, Ruiter JPN, IJlst L, Duran M, et al. Valproic acid metabolism and its effects on mitochondrial fatty acid oxidation: A review. J Inherit Metab Dis. 2008;31:205–216. doi: 10.1007/s10545-008-0841-x. [DOI] [PubMed] [Google Scholar]
- 11.Silva MFB, Ruiter JP, Illst L, Jakobs C, Duran M, de Almeida IT, Wanders RJ. Valproate inhibits the mitochondrial pyruvate-driven oxidative phosphorylation in vitro. J Inherit Metab Dis. 1997;20:397–400. doi: 10.1023/a:1005398516208. [DOI] [PubMed] [Google Scholar]
- 12.Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;336:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 13.Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bender T, Martinou JC. The mitochondrial pyruvate carrier in health and disease: to carry or not to carry? Biochim Biophys Acta. 2016;1863:2436–2442. doi: 10.1016/j.bbamcr.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 15.Rauckhorst AJ, Taylor EB. Mitochondrial pyruvate carrier function and cancer metabolism. Curr Opin Genet Dev. 2016;38:102–109. doi: 10.1016/j.gde.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensard CL, Wisidagama DR, Olson KA, Berg JA, Krah NM, Schell JC, Nowinski SM, Fogarty S, Bott AJ, Wei P, et al. Regulation of tumor initiation by the mitochondrial pyruvate carrier. Cell Metab. 2020;31:284–300. doi: 10.1016/j.cmet.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Taufalele PV, Cochran JD, Robillard-Frayne I, Marx JM, Soto J, Rauckhorst AJ, Tayyari F, Pewa AD, Gray LR, Teesch LM, Puchalska P, Funari TR, McGlauflin R, Zimmerman K, Kutschke WJ, Cassier T, Hitchcock S, Lin K, Kato KM, Stueve JL, Haff L, Weiss RM, Cox JE, Rutter J, Taylor EB, Crawford PA, Lewandowski ED, Des Rosiers C, Abel ED. Mitochondrial pyruvate carriers are required for myocardial stress adaptation. Nat Metab. 2020;2(11):1248–1264. doi: 10.1038/s42255-020-00288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh A, Tyson T, George S, Hildebrandt EN, Steiner JA, Madaj Z, Schulz E, Machiela E, McDonald WG, Escobar Galvis ML, Kordower JH, Van Raamsdonk JM, Colca JR, Brundin P. Mitochondrial pyruvate carrier regulates autophagy, inflammation, and neurodegeneration in experimental models of Parkinson's disease. Sci Transl Med. 2016;8(368):36ra8174. doi: 10.1126/scitranslmed.aag2210. [DOI] [PubMed] [Google Scholar]
- 19.Rossi A, Rigotto G, Valente G, Giorgio V, Basso E, Filadi R, Pizzo P. Defective mitochondrial pyruvate flux affects cell bioenergetics in Alzheimer’s disease-related models. Cell Rep. 2020;30:2332–2348. doi: 10.1016/j.celrep.2020.01.060. [DOI] [PubMed] [Google Scholar]
- 20.Jeoung NH, Harris CR, Harris RA. Regulation of pyruvate metabolism in metabolic-related diseases. Rev Endocr Metab Disord. 2014;15:99–110. doi: 10.1007/s11154-013-9284-2. [DOI] [PubMed] [Google Scholar]
- 21.Zangari J, Petrelli F, Maillot B, Martinou JC. The multifaceted pyruvate metabolism: role of the mitochondrial pyruvate carrier. Biomolecules. 2020;10:1068. doi: 10.3390/biom10071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCommis KS, Chen Z, Fu X, McDonald WG, Colca JR, Kletzien RF, Burgess SC, Finck BN. Loss of mitochondrial pyruvate carrier 2 in liver leads to defects in gluconeogenesis and compensation via pyruvate-alanine cycling. Cell Metab. 2015;22:682–694. doi: 10.1016/j.cmet.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hezaveh S, Zeng AP, Jandt U. Full enzyme complex simulation: interactions in human pyruvate dehydrogenase complex. J Chem Inf Model. 2018;58:362–369. doi: 10.1021/acs.jcim.7b00557. [DOI] [PubMed] [Google Scholar]
- 24.Harris RA, Bowker-Kinley MM, Wu P, Jeng J, Popov KM. Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex. DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J Biol Chem. 1997;272(32):19746–19751. doi: 10.1074/jbc.272.32.19746. [DOI] [PubMed] [Google Scholar]
- 25.Kato M, Wynn RM, Chuang JL, Tso SC, Machius M, Li J, Chuang DT. Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops. Structure. 2008;16(12):1849–1859. doi: 10.1016/j.str.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhandary S, Aguan K. Pyruvate dehydrogenase complex deficiency and its relationship with epilepsy frequency—An overview. Epilepsy Res. 2015;116:40–52. doi: 10.1016/j.eplepsyres.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Song J, Yan L-J. Chronic inhibition of mitochondrial dihydrolipoamide dehydrogenase (DLD) as an approach to managing diabetic oxidative stress. Antioxidants. 2019;8:32. doi: 10.3390/antiox8020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan L-J, Thangthaeng N, Sumien N, Forster MJ. Serum dihydrolipoamide dehydrogenase is a labile enzyme. J Biochem Pharmacol Res. 2013;1:30–42. [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Luo X, Wu J, Thangthaeng N, Jung ME, Jing S, et al. Mitochondrial dihydrolipoamide dehydrogenase is upregulated in response to intermittent hypoxic preconditioning. Int J Med Sci. 2015;12:432–440. doi: 10.7150/ijms.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, Li J, Guo Z, Sun L, Juan C, Zhou Y, et al. Overexpression of pyruvate dehydrogenase e1a subunit inhibits warburg effect and induces cell apoptosis through mitochondria-mediated pathway in hepatocellular carcinoma. Oncol Res. 2019;27:407–414. doi: 10.3727/096504018X15180451872087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonashiro R, Eguchi K, Wake M, Takeda N, Nakayama K. Pyruvate dehydrogenase PDH-E1b controls tumor progression by altering the metabolic status of cancer cells. Cancer Res. 2018;78:1592–1603. doi: 10.1158/0008-5472.CAN-17-1751. [DOI] [PubMed] [Google Scholar]
- 32.Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 2003;31(6):1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- 33.Linn TC, Pettit FH, Reed LJ. Alpha-keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc Natl Acad Sci USA. 1969;62:234–241. doi: 10.1073/pnas.62.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeoung NH. Pyruvate dehydrogenase kinases: therapeutic targets for diabetes and cancers. Diabetes Metab J. 2015;39:188–197. doi: 10.4093/dmj.2015.39.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eguchi K, Nakayama K. Prolonged hypoxia decreases nuclear pyruvate dehydrogenase complex and regulates the gene expression. Biochem Biophys Res Commun. 2019;520:128–135. doi: 10.1016/j.bbrc.2019.09.109. [DOI] [PubMed] [Google Scholar]
- 36.Wigfield SM, Winter SC, Giatromanolaki A, Taylor J, Koukourakis ML, Harris AL. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br J Cancer. 2008;98:1975–1984. doi: 10.1038/sj.bjc.6604356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyassu F, Angione C. Modelling pyruvate dehydrogenase under hypoxia and its role in cancer metabolism. R Soc Open Sci. 2017;4:170360. doi: 10.1098/rsos.170360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. JNCI J Natl Cancer Inst. 2017;109:1–14. doi: 10.1093/jnci/djx071. [DOI] [PubMed] [Google Scholar]
- 39.Nasiri A, Sadeghi M, Vaisi-Raygani A, Kiani S, Aghelan Z, Khodarahmi R. Emerging regulatory roles of mitochondrial sirtuins on pyruvate dehydrogenase complex and the related metabolic diseases: review. Biomed Res Ther. 2020;7:3645–3658. [Google Scholar]
- 40.Park JM, Reed GD, Liticker J, Putnam WC, Chandra A, Yaros K, Afzal A, MacNamara J, Raza J, Hall RG, Baxter J, Derner K, Pena S, Kallem RR, Subramaniyan I, Edpuganti V, Harrison CE, Muthukumar A, Lewis C, Reddy S, Unni N, Klemow D, Syed S, Li H, Cole S, Froehlich T, Ayers C, de Lemos J, Malloy CR, Haley B, Zaha VG. Effect of doxorubicin on myocardial bicarbonate production from pyruvate dehydrogenase in women with breast cancer. Circ Res. 2020;127(12):1568–1570. doi: 10.1161/CIRCRESAHA.120.317970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang SL, Hu X, Zhang W, Yao H, Tam KY. Development of pyruvate dehydrogenase kinase inhibitors in medicinal chemistry with particular emphasis as anticancer agents. Drug Discov Today. 2015;20(9):1112–1119. doi: 10.1016/j.drudis.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Pavlu-Pereira H, Silva MJ, Florindo C, Sequeira S, Ferreira AC, Duarte S, Rodrigues AL, Janeiro P, Oliveira A, Gomes D, Bandeira A, Martins E, Gomes R, Soares S, Tavares de Almeida I, Vicente JB, Rivera I. Pyruvate dehydrogenase complex deficiency: updating the clinical, metabolic and mutational landscapes in a cohort of Portuguese patients. Orphanet J Rare Dis. 2020;15(1):298. doi: 10.1186/s13023-020-01586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrus A, Adam-Vizi V. Human dihydrolipoamide dehydrogenase (E3) deficiency: novel insights into the structural basis and molecular pathomechanism. Neurochem Int. 2018;117:5–14. doi: 10.1016/j.neuint.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 44.Babady NE, Pang YP, Elpeleg O, Isaya G. Cryptic proteolytic activity of dihydrolipoamide dehydrogenase. Proc Natl Acad Sci USA. 2007;104:6158–6163. doi: 10.1073/pnas.0610618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dayan A, Yeheskel A, Lamed R, Fleminger G, Ashur-Fabian O (2020) Dihydrolipoamide dehydrogenase moonlighting activity as a DNA chelating agent. Proteins: 1–8. 10.1002/prot.25991 [DOI] [PubMed]
- 46.Ciszak EM, Korotchkina LG, Dominiak PM, Sidhu S, Patel MS. Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase. J Biol Chem. 2003;278(23):21240–21246. doi: 10.1074/jbc.M300339200. [DOI] [PubMed] [Google Scholar]
- 47.Ciszak EM, Makal A, Hong YS, Vettaikkorumakankauv AK, Korotchkina LG, Patel MS. How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex. J Biol Chem. 2006;281(1):648–655. doi: 10.1074/jbc.M507850200. [DOI] [PubMed] [Google Scholar]
- 48.Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J Mol Biol. 2005;350:543–552. doi: 10.1016/j.jmb.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Brautigam CA, Wynn RM, Chuang JL, Machius M, Tomchick DR, Chuang DT. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure. 2006;14:611–621. doi: 10.1016/j.str.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brautigam CA, Wynn RM, Chuang JL, Chuang DT. Subunit and catalytic component stoichiometries of an in vitro reconstituted human pyruvate dehydrogenase complex. J Biol Chem. 2009;284(19):13086–13098. doi: 10.1074/jbc.M806563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brautigam CA, Wynn RM, Chuang JL, Naik MT, Young BB, Huang TH, Chuang DT. Structural and thermodynamic basis for weak interactions between dihydrolipoamide dehydrogenase and subunit-binding domain of the branched-chain alpha-ketoacid dehydrogenase complex. J Biol Chem. 2011;286:23476–23488. doi: 10.1074/jbc.M110.202960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szabo E, Mizsei R, Wilk P, Zambo Z, Torocsik B, Weiss MS, Adam-Vizi V, Ambrus A. Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase. Free Radic Biol Med. 2018;124:214–220. doi: 10.1016/j.freeradbiomed.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Szabo E, Wilk P, Nagy B, Zambo Z, Bui D, Weichsel A, Arjunan P, Torocsik B, Hubert A, Furey W, Montfort WR, Jordan F, Weiss MS, Adam-Vizi V, Ambrus A. Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants. Hum Mol Genet. 2019;28(20):3339–3354. doi: 10.1093/hmg/ddz177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klyachko NL, Shchedrina VA, Efimov AV, Kazakov SV, Gazaryan IG, Kristal BS, Brown AM. pH-dependent substrate preference of pig heart lipoamide dehydrogenase varies with oligomeric state: response to mitochondrial matrix acidification. J Biol Chem. 2005;280(16):16106–16114. doi: 10.1074/jbc.M414285200. [DOI] [PubMed] [Google Scholar]
- 55.Ambrus A. An updated view on the molecular pathomechanisms of human dihydrolipoamide dehydrogenase deficiency in light of novel crystallographic evidence. Neurochem Res. 2019;44:2307–2313. doi: 10.1007/s11064-019-02766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nemeria NS, Gerfen G, Nareddy PR, Yang L, Zhang X, Szostak M, et al. The mitochondrial 2-oxoadipate and 2-oxoglutarate dehydrogenase complexes share their E2 and E3 components for their function and both generate reactive oxygen species. Free Radic Biol Med. 2018;115:136–145. doi: 10.1016/j.freeradbiomed.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 57.Bezerra GA, Foster WR, Bailey HJ, Hicks KG, Sauer SW, Dimitrov B, et al. Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism. IUCrJ. 2020;7:693–706. doi: 10.1107/S205225252000696X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The UniProt Consortium (2018) UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res 46(5):2699. https://www.uniprot.org/uniprot/P09622. Assessed Sept 2020 [DOI] [PMC free article] [PubMed]
- 59.Fukamichi T, Nishimoto E. Conformational change near the redox center of dihydrolipoamide dehydrogenase induced by NAD+ to regulate the enzyme activity. J Fluoresc. 2015;25:577–583. doi: 10.1007/s10895-015-1537-x. [DOI] [PubMed] [Google Scholar]
- 60.Yan L-J, Liu L, Forster MJ. Reversible inactivation of dihydrolipoamide dehydrogenase by Angeli’s salt. Sheng Wu Wu Li Hsueh Bao. 2012;28:341–350. [PMC free article] [PubMed] [Google Scholar]
- 61.Velankar S, Kleywegt GJ (2011) The protein data bank in europe (PDBe): bringing structure to biology. Acta Crystallogr D 67:234–330. https://www.ebi.ac.uk/pdbe/. Accessed 7 Sept 2020 [DOI] [PMC free article] [PubMed]
- 62.Quinonez SC, Thoene JG. Dihydrolipoamide dehydrogenase deficiency. 2014 Jul 17 [Updated 2020 Jul 9] In: Adam MP, Ardinger HH, Pagon RA, editors. GeneReviews® [Internet] Seattle (WA): University of Washington; 2014. pp. 1993–2021. [PubMed] [Google Scholar]
- 63.Cameron JM, Levandovskiy V, Mackay N, Raiman J, Renaud DL, Clarke JTR, Feigenbaum A, Elpeleg O, Robison BH. Novel mutations in dihydrolipoamide dehydrogenase deficiency in two cousins with borderline-normal PDH complex activity. Am J Med Genet A. 2006;140:1542–1552. doi: 10.1002/ajmg.a.31313. [DOI] [PubMed] [Google Scholar]
- 64.Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. 2014;289(12):8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Igamberdiev AU, Bykova NV, Ens W, Hill RD. Dihydrolipoamide dehydrogenase from porcine heart catalyzes NADH-dependent scavenging of nitric oxide. FEBS Lett. 2004;568:146–150. doi: 10.1016/j.febslet.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 66.Ambrus A, Torocsik B, Tretter L, Ozohanics O, Adam-Vizi V. Stimulation of reactive oxygen species generation by disease-causing mutations of lipoamide dehydrogenase. Hum Mol Genet. 2011;20:2984–2995. doi: 10.1093/hmg/ddr202. [DOI] [PubMed] [Google Scholar]
- 67.Ambrus A, Adam-Vizi V. Molecular dynamics study of the structural basis of dysfunction and the modulation of reactive oxygen species generation by pathogenic mutants of human dihydrolipoamide dehydrogenase. Arch Biochem Biophys. 2013;538(2):145–155. doi: 10.1016/j.abb.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 68.Ambrus A, Nemeria NS, Torocsika B, Tretter L, Nilsson M, Jordan F, Adam-Vizi V. Formation of reactive oxygen species by human and bacterial pyruvate and 2-oxoglutarate dehydrogenase multienzyme complexes reconstituted from recombinant components. Free Radic Biol Med. 2015;89:642–650. doi: 10.1016/j.freeradbiomed.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ambrus A, Wang J, Mizsei R, Zambo Z, Torocsik B, Jordan F, Adam-Vizi V. Structural alterations induced by ten disease-causing mutations of human dihydrolipoamide dehydrogenase analyzed by hydrogen/deuterium-exchange mass spectrometry: implications for the structural basis of E3 deficiency. Biochim Biophys Acta. 2016;1862:2098–2109. doi: 10.1016/j.bbadis.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mailloux RJ, Gardiner D, O’Brien M. 2-Oxoglutarate dehydrogenase is a more significant source of O2.-/H2O2 than pyruvate dehydrogenase in cardiac and liver tissue. Free Radic Biol Med. 2016;97:501–512. doi: 10.1016/j.freeradbiomed.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 71.Yan LJ, Thangthaeng N, Forster MJ. Changes in dihydrolipoamide dehydrogenase expression and activity during postnatal development and aging in the rat brain. Mech Ageing Dev. 2008;129:282–290. doi: 10.1016/j.mad.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin D, Lee J, You JH, Kim D, Roh JL. Dihydrolipoamide dehydrogenase regulates cystine deprivation-induced ferroptosis in head and neck cancer. Redox Biol. 2020;30:101418. doi: 10.1016/j.redox.2019.101418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dayan A, Fleminger G, Ashur-Fabian O. Targeting the Achilles’ heel of cancer cells via integrin-mediated delivery of ROS-generating dihydrolipoamide dehydrogenase. Oncogene. 2019;38(25):5050–5061. doi: 10.1038/s41388-019-0775-9. [DOI] [PubMed] [Google Scholar]
- 74.Olson KA, Schell JC, Rutter J. Pyruvate and metabolic flexibility: illuminating a path toward selective cancer therapies. Trends Biochem Sci. 2016;41(3):219–230. doi: 10.1016/j.tibs.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaimes R, 3rd, Kuzmiak-Glancy S, Brooks DM, Swift LM, Posnack NG, Kay MW. Functional response of the isolated, perfused normoxic heart to pyruvate dehydrogenase activation by dichloroacetate and pyruvate. Pflugers Arch. 2016;468:131–142. doi: 10.1007/s00424-015-1717-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gazaryan IG, Krasnikov BF, Ashby GA, Thorneley RNF, Kristal BS, Brown AM. Zinc is a potent inhibitor of thiol oxidoreductase activity and stimulates reactive oxygen species production by lipoamide dehydrogenase. J Biol Chem. 2002;277(12):10064–10072. doi: 10.1074/jbc.M108264200. [DOI] [PubMed] [Google Scholar]
- 77.Gazaryan IG, Shchedrina VA, Klyachko NL, Zakhariants AA, Kazakov SV, Brown AM. Zinc switch in pig heart lipoamide dehydrogenase: steady-state and transient kinetic studies of the diaphorase reaction. Biochemistry (Mosc) 2020;85(8):908–919. doi: 10.1134/S0006297920080064. [DOI] [PubMed] [Google Scholar]
- 78.Schlipalius DI, Valmas N, Tuck AG, Jagadeesan R, Ma L, Kaur R, Goldinger A, Anderson C, Kuang J, Zuryn S, Mau YS, Cheng Q, Collins PJ, Nayak MK, Schirra HJ, Hilliard MA, Ebert PR. A core metabolic enzyme mediates resistance to phosphine gas. Science. 2012;338:807–810. doi: 10.1126/science.1224951. [DOI] [PubMed] [Google Scholar]
- 79.Alzahrani S, Ebert PR. Oxygen and arsenite synergize phosphine toxicity by distinct mechanisms. Toxicol Sci. 2019;167(2):419–425. doi: 10.1093/toxsci/kfy248. [DOI] [PubMed] [Google Scholar]
- 80.Yan LJ. Reexploring 5-methoxyindole-2-carboxylic acid (MICA) as a potential antidiabetic agent. Diabetes Metab Syndr Obes. 2018;11:183–186. doi: 10.2147/DMSO.S166485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luís PBM, Ruiter JPN, Aires CCP, Soveral G, Tavares de Almeida I, Duran M, et al. Valproic acid metabolites inhibit dihydrolipoyl dehydrogenase activity leading to impaired 2-oxoglutarate-driven oxidative phosphorylation. Biochim Biophys Acta. 2007;1767:1126–1133. doi: 10.1016/j.bbabio.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 82.Tomson T, Battino D, Perucca E. Valproic acid after five decades of use in epilepsy: time to reconsider the indications of a time-honoured drug. Lancet Neurol. 2016;15:210–218. doi: 10.1016/S1474-4422(15)00314-2. [DOI] [PubMed] [Google Scholar]
- 83.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 84.Jeong MR, Hashimoto R, Senatorov VV, Fujimaki K, Ren M, Lee MS, et al. Valproic acid, a mood stabilizer and anticonvulsant, protects rat cerebral cortical neurons from spontaneous cell death: a role of histone deacetylase inhibition. FEBS Lett. 2003;542:74–78. doi: 10.1016/s0014-5793(03)00350-8. [DOI] [PubMed] [Google Scholar]
- 85.Dufour-Rainfray D, Vourc’h P, Tourlet S, Guilloteau D, Chalon S, Andres CR. Fetal exposure to teratogens: evidence of genes involved in autism. Neurosci Biobehav Rev. 2011;35:1254–1265. doi: 10.1016/j.neubiorev.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 86.Evers S, Áfra J, Frese A, Goadsby PJ, Linde M, May A, et al. EFNS guideline on the drug treatment of migraine—Revised report of an EFNS task force. Eur J Neurol. 2009;16:968–981. doi: 10.1111/j.1468-1331.2009.02748.x. [DOI] [PubMed] [Google Scholar]
- 87.Steiner TJ, Jensen R, Katsarava Z, Linde M, MacGregor EA, Osipova V, et al. Aids to management of headache disorders in primary care (2nd edition): on behalf of the European Headache Federation and Lifting the Burden: The Global Campaign against Headache. J Headache Pain. 2019;20:57. doi: 10.1186/s10194-018-0899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lloyd KA. A scientific review: mechanisms of valproate-mediated teratogenesis. Biosci Horiz. 2013;6:1–10. [Google Scholar]
- 89.Chen Y, Zhou J, Xu S, Liu M, Wang M, Ma Y, Zhao M, Wang Z, Guo Y, Zhao L. Association between the perturbation of bile acid homeostasis and valproic acid-induced hepatotoxicity. Biochem Pharmacol. 2019;170:113669. doi: 10.1016/j.bcp.2019.113669. [DOI] [PubMed] [Google Scholar]
- 90.Xu S, Chen Y, Ma Y, Liu T, Zhao M, Wang Z, et al. Lipidomic profiling reveals disruption of lipid metabolism in valproic acid-induced hepatotoxicity. Front Pharmacol. 2019;10:819. doi: 10.3389/fphar.2019.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clayton-Smith J, Bromley R, Dean J, Journel H, Odent S, Wood A, et al. Diagnosis and management of individuals with fetal valproate spectrum disorder; a consensus statement from the European reference network for congenital malformations and intellectual disability. Orphanet J Rare Dis. 2019;14:180. doi: 10.1186/s13023-019-1064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nanau RM, Neuman MG. Adverse drug reactions induced by valproic acid. Clin Biochem. 2013;46:1323–1338. doi: 10.1016/j.clinbiochem.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 93.Gopaul S, Farrell K, Abbott F. Effects of age and polytherapy, risk factors of valproic acid (VPA) hepatotoxicity, on the excretion of thiol conjugates of (E)-2,4-diene VPA in people with epilepsy taking VPA. Epilepsia. 2003;44:322–328. doi: 10.1046/j.1528-1157.2003.07202.x. [DOI] [PubMed] [Google Scholar]
- 94.Begriche K, Massart J, Robin MA, Sanchez AB, Fromenty B. Drug-induced toxicity on mitochondria and lipid metabolism: mechanistic diversity and deleterious consequences for the liver. J Hepatol. 2011;54:773–794. doi: 10.1016/j.jhep.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 95.LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet]. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases; 2012-.Valproate. (Updated 2020, July 31). https://www.ncbi.nlm.nih.gov/books/NBK547852/. Accessed 10 Sept 2020 [PubMed]
- 96.Felker D, Lynn A, Wang S, Johnson DE. Evidence for a potential protective effect of carnitine-pantothenic acid co-treatment on valproic acid-induced hepatotoxicity. Expert Rev Clin Pharmacol. 2014;7:211–218. doi: 10.1586/17512433.2014.871202. [DOI] [PubMed] [Google Scholar]
- 97.Lheureux PER, Hantson P. Carnitine in the treatment of valproic acid-induced toxicity. Clin Toxicol. 2009;47:101–111. doi: 10.1080/15563650902752376. [DOI] [PubMed] [Google Scholar]
- 98.Abdelkader NF, Elyamany M, Gad AM, Assaf N, Fawzy HM, Elesawy WH. Ellagic acid attenuates liver toxicity induced by valproic acid in rats. J Pharmacol Sci. 2020;143:23–29. doi: 10.1016/j.jphs.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 99.Oztopuz O, Turkon H, Buyuk B, Coskun O, Sehitoglu MH, Ovali MA, et al. Melatonin ameliorates sodium valproate-induced hepatotoxicity in rats. Mol Biol Rep. 2020;47:317–325. doi: 10.1007/s11033-019-05134-6. [DOI] [PubMed] [Google Scholar]
- 100.Silva MFB, Ruiter JPN, Overmars H, Bootsma AH, van Gennip AH, Jakobs C, Duran M, de Almeida IT, Wanders RJ. Complete β-oxidation of valproate: cleavage of 3-oxovalproyl-CoA by a mitochondrial 3-oxoacyl-CoA thiolase. Biochem J. 2002;362:755–760. doi: 10.1042/0264-6021:3620755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aires CCP, Ruiter JPN, Luís PBM, ten Brink HJ, IJlst L, de Almeida IT, Duran M, Wanders RJA, Silva MFB. Studies on the extra-mitochondrial CoA-ester formation of valproic and Δ4-valproic acids. Biochim Biophys Acta. 2007;1771:533–543. doi: 10.1016/j.bbalip.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 102.Silva MFB, Ruiter JPN, IJlst L, Allers P, Ten Brink HJ, Jakobs C, Duran M, de Almeida IT, Wanders RJ. Synthesis and intramitochondrial levels of valproyl-coenzyme A metabolites. Anal Biochem. 2001;290:60–67. doi: 10.1006/abio.2000.4947. [DOI] [PubMed] [Google Scholar]
- 103.Luís PBM, Ruiter JP, Ofman R, Ijlst L, Moedas M, Diogo L, Garcia P, de Almeida IT, Duran M, Wanders RJA, Silva MFB. Valproic acid utilizes the isoleucine breakdown pathway for its complete β-oxidation. Biochem Pharmacol. 2011;82:1740–1746. doi: 10.1016/j.bcp.2011.07.103. [DOI] [PubMed] [Google Scholar]
- 104.Luís PBM, Ruiter JPN, IJlst L, de Almeida IT, Duran M, Mohsen A, Vockley J, Wanders RJ, Silva MFB. Role of isovaleryl-CoA dehydrogenase and short branched-chain acyl-CoA dehydrogenase in the metabolism of valproic acid: implications for the branched-chain amino acid oxidation pathway. Drug Metab Dispos. 2011;39:1155–1160. doi: 10.1124/dmd.110.037606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aires CC, Soveral G, Luís PBM, ten Brink HJ, de Almeida IT, Duran M, Wanders RJA, Silva MFB. Pyruvate uptake is inhibited by valproic acid and metabolites in mitochondrial membranes. FEBS Lett. 2008;582:3359–3366. doi: 10.1016/j.febslet.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 106.Benavides J, Martin A, Ugarte M, Valdivieso F. Inhibition by valproic acid of pyruvate uptake by brain mitochondria. Biochem Pharmacol. 1982;31(8):1633–1636. doi: 10.1016/0006-2952(82)90392-6. [DOI] [PubMed] [Google Scholar]
- 107.Kudin AP, Mawasi H, Eisenkraft A, Elger CE, Bialer M, Kunz WS. Mitochondrial liver toxicity of valproic acid and its acid derivatives is related to inhibition of α-lipoamide dehydrogenase. Int J Mol Sci. 2017;18:1912–1923. doi: 10.3390/ijms18091912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deutsch J, Rapoport SI, Rosenberger TA. Valproyl-CoA and esterified valproic acid are not found in brains of rats treated with valproic acid, but the brain concentrations of CoA and acetyl-CoA are altered. Neurochem Res. 2003;28:861–866. doi: 10.1023/a:1023267224819. [DOI] [PubMed] [Google Scholar]
- 109.Mampilly GT, Mampilly TK, Christopher R, Chandramohan N, Janaki V. Challenges in diagnosing a metabolic disorder: error of pyruvate metabolism or drug induced? J Child Neurol. 2014;29(6):833–836. doi: 10.1177/0883073813477201. [DOI] [PubMed] [Google Scholar]
- 110.Moedas MF, van Cruchten AG, IJlst L, Kulik W, de Almeida IT, Diogo L, Wanders RJ, Silva MFB. Transient decrease of hepatic NAD+ and amino acid alterations during treatment with valproate: new insights on drug-induced effects in vivo using targeted MS-based metabolomics. Metabolomics. 2016;12:142. [Google Scholar]
- 111.Huo T, Chen X, Lud X, Qub L, Liub Y, Cai S. An effective assessment of valproate sodium-induced hepatotoxicity with UPLC–MS and 1HNMR-based metabonomics approach. J Chromatogr B. 2014;969:109–116. doi: 10.1016/j.jchromb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 112.Mortensen PB, Kplvraa S, Christensen E. Inhibition of the glycine cleavage system: hyperglycinemia and hyperglycinuria caused by valproic acid. Epilepsia. 1980;21:563–569. doi: 10.1111/j.1528-1157.1980.tb04310.x. [DOI] [PubMed] [Google Scholar]
- 113.Martin-Gallardo A, Rodriguez P, Lopez M, Benavides J, Ugarte M. Effects of dipropylacetate on the glycine cleavage enzyme system and glycine levels. A possible experimental approach to non-ketotic hyperglycinemia. Biochem Pharmacol. 1985;34:2877–2882. doi: 10.1016/0006-2952(85)90010-3. [DOI] [PubMed] [Google Scholar]
- 114.Yoshino M, Koga Y, Yamashita F. A decrease in glycine cleavage activity in the liver of a patient with dihydrolipoyl dehydrogenase deficiency. J Inher Metab Dis. 1986;9:399–400. doi: 10.1007/BF01800493. [DOI] [PubMed] [Google Scholar]
- 115.Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W. Lipoic acid biosynthesis defects. J Inher Metab Dis. 2014;37:553–563. doi: 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 116.Tort F, Ferrer-Cortes X, Ribes A. Differential diagnosis of lipoic acid synthesis defects. J Inher Metab Dis. 2016;39:781–793. doi: 10.1007/s10545-016-9975-4. [DOI] [PubMed] [Google Scholar]
- 117.Cronan JE. Progress in the enzymology of the mitochondrial diseases of lipoic acid requiring enzymes. Front Genet. 2020;11:510. doi: 10.3389/fgene.2020.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jia F, Cui M, Than MT, Han M. Developmental defects of Caenorhabditis elegans lacking branched-chain α-ketoacid dehydrogenase are mainly caused by monomethyl branched-chain fatty acid deficiency. J Biol Chem. 2015;291:2967–2973. doi: 10.1074/jbc.M115.676650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heinemann-Yerushalmi L, Bentovim L, Felsenthal N, Michaeli N, Krief S, Haffner-Krausz R, et al. BCKDK regulates the TCA cycle through PDC in the absence of PDK family during embryonic development. Dev Cell. 2021;56(8):1182–1194.e6. doi: 10.1016/j.devcel.2021.03.007. [DOI] [PubMed] [Google Scholar]
- 120.Luís PB, Ruiter JP, IJlst L, Diogo L, Garcia P, de Almeida IT, Duran M, Wanders RJ, Silva MFB. Inhibition of 3-methylcrotonyl-CoA carboxylase explains the increased excretion of 3-hydroxyisovaleric acid in valproate-treated patients. J Inherit Metab Dis. 2012;35(3):443–449. doi: 10.1007/s10545-011-9423-4. [DOI] [PubMed] [Google Scholar]
- 121.Maciejak P, Szyndler J, Kołosowska K, Turzyńska D, Sobolewska A, Walkowiak J, Płaźnik A. Valproate disturbs the balance between branched and aromatic amino acids in rats. Neurotox Res. 2014;25:358–368. doi: 10.1007/s12640-013-9441-0. [DOI] [PubMed] [Google Scholar]
- 122.Ævarsson A, Chuang JL, Wynn RM, Turley S, Chuang DT, Hol WG. Crystal structure of human branched-chain α-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure. 2000;8:277–291. doi: 10.1016/s0969-2126(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 123.Zhou J, Yang L, Ozohanics O, Zhang X, Wang J, Ambrus A, Arjunan P, Brukh R, Nemeria NS, Furey W, Jordan F. A multipronged approach unravels unprecedented protein-protein interactions in the human 2-oxoglutarate dehydrogenase multienzyme complex. J Biol Chem. 2018;293(50):19213–19227. doi: 10.1074/jbc.RA118.005432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luder AS, Parks JK, Frerman F, Parker WD. Inactivation of beef brain α-ketoglutarate dehydrogenase complex by valproic acid and valproic acid metabolites. Possible mechanism of anticonsulvant and toxic actions. J Clin Invest. 1990;86:1574–1581. doi: 10.1172/JCI114877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piplani S, Verma PK, Kumar A. Neuroinformatics analyses reveal GABAt and SSADH as major proteins involved in anticonvulsant activity of valproic acid. Biomed Pharmacother. 2016;81:402–410. doi: 10.1016/j.biopha.2016.04.036. [DOI] [PubMed] [Google Scholar]
- 126.Dobolyi A, Bago A, Palkovits M, Nemeria NS, Jordan F, Doczi J, Ambrus A, Adam-Vizi V, Chinopoulos C. Exclusive neuronal detection of KGDHC-specific subunits in the adult human brain cortex despite pancellular protein lysine succinylation. Brain Struct Funct. 2020;225:639–667. doi: 10.1007/s00429-020-02026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Phil Trans R Soc B. 2005;360:2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moxley MA, Beard DA, Bazil JN. Global Kinetic Analysis of mammalian E3 reveals pH-dependent NAD+/NADH regulation, physiological kinetic reversibility and catalytic optimum. J Biol Chem. 2016;291(6):2712–2730. doi: 10.1074/jbc.M115.676619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shibata K, Kondo R, Sano M, Fukuwatari T. Increased conversion of tryptophan to nicotinamide in rats by dietary valproate. Biosci Biotechnol Biochem. 2013;77:295–300. doi: 10.1271/bbb.120716. [DOI] [PubMed] [Google Scholar]
- 130.Klimova N, Fearnow A, Kristian T. Role of NAD+-modulated mitochondrial free radical generation in mechanisms of acute brain injury. Brain Sci. 2020;10:449. doi: 10.3390/brainsci10070449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang X, Wang A, Zhu L, Hua D, Qin J. Altering the sensitivity of Escherichia coli pyruvate dehydrogenase complex to NADH inhibition by structure-guided design. Enzyme Microb Technol. 2018;119:52–57. doi: 10.1016/j.enzmictec.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 132.Solmonson A, DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem. 2018;293:7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nemeria NS, Zhang X, Leandro J, Zhou J, Yang L, Houten SM, Jordan F. Toward an understanding of the structural and mechanistic aspects of protein-protein interactions in 2-oxoacid dehydrogenase complexes. Life (Basel) 2021;11(5):407. doi: 10.3390/life11050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16(4):258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 136.Park S, Jeon JH, Min BK, Ha CM, Thoudam T, Park BY, Lee IK. Role of the pyruvate dehydrogenase complex in metabolic remodeling: differential pyruvate dehydrogenase complex functions in metabolism. Diabetes Metab J. 2018;42:270–281. doi: 10.4093/dmj.2018.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ali I, Conrad RJ, Verdin E, Ott M. Lysine acetylation goes global: from epigenetics to metabolism and therapeutics. Chem Rev. 2018;118:1216–1252. doi: 10.1021/acs.chemrev.7b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 139.Ye C, Tu BP. Sink into the Epigenome: histones as repositories that influence cellular metabolism. Trends Endocrinol Metab. 2018;29:626–637. doi: 10.1016/j.tem.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10:95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sivanand S, Viney I, Wellen KE. Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends Biochem Sci. 2018;43:61–74. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ng F, Tang BL. Pyruvate dehydrogenase complex (PDC) export from the mitochondrial matrix. Mol Membr Biol. 2014;31(7–8):207–210. doi: 10.3109/09687688.2014.987183. [DOI] [PubMed] [Google Scholar]
- 144.Narne P, Pandey V, Phanithi PB. Interplay between mitochondrial metabolism and oxidative stress in ischemic stroke: an epigenetic connection. Mol Cell Neurosci. 2017;82:176–194. doi: 10.1016/j.mcn.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 145.Martínez-Reyes I, Chandel NS. Acetyl-CoA-directed gene transcription in cancer cells. Genes Dev. 2018;32:463–465. doi: 10.1101/gad.315168.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 147.Itoh Y. Drug discovery researches on modulators of lysine-modifying enzymes based on strategic chemistry approaches. Chem Pharm Bull. 2020;68:34–45. doi: 10.1248/cpb.c19-00741. [DOI] [PubMed] [Google Scholar]
- 148.Gil J, Ramírez-Torres A, Encarnación-Guevara S. Lysine acetylation and cancer: a proteomics perspective. J Proteom. 2017;150:297–309. doi: 10.1016/j.jprot.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 149.Li P, Ge J, Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat Rev Cardiol. 2020;17:96–115. doi: 10.1038/s41569-019-0235-9. [DOI] [PubMed] [Google Scholar]
- 150.Göttlicher M, Minucci S, Zhu P, Krämer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20(24):6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van Breda SGJ, Claessen SMH, van Herwijnen M, Theunissen DHJ, Jennen DGJ, de Kok TMCM, et al. Integrative omics data analyses of repeated dose toxicity of valproic acid in vitro reveal new mechanisms of steatosis induction. Toxicology. 2018;393:160–170. doi: 10.1016/j.tox.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 152.Blaheta RA, Michaelis M, Driever PH, Cinatl J. Evolving anticancer drug valproic acid: insights into the mechanism and clinical studies. Med Res Rev. 2005;25:383–397. doi: 10.1002/med.20027. [DOI] [PubMed] [Google Scholar]
- 153.Yu J, Choi C, Shin SW, Son A, Lee GH, Kim SY, Park HC. Valproic acid sensitizes hepatocellular carcinoma cells to proton therapy by suppressing NRF2 activation. Sci Rep. 2017;7:14986. doi: 10.1038/s41598-017-15165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ibrahim TS, Sheha TA, Abo-Dya NE, AlAwadh MA, Alhakamy NA, Abdel-Samii ZK, et al. Design, synthesis and anticancer activity of novel valproic acid conjugates with improved histone deacetylase (HDAC) inhibitory activity. Bioorg Chem. 2020;99:103797. doi: 10.1016/j.bioorg.2020.103797. [DOI] [PubMed] [Google Scholar]
- 155.Lu P, Yan M, He L, Li J, Ji Y, Ji J. Crosstalk between epigenetic modulations in valproic acid deactivated hepatic stellate cells: an integrated protein and miRNA profiling study. Int J Biol Sci. 2019;15:93–104. doi: 10.7150/ijbs.28642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Georgoff PE, Halaweish I, Nikolian VC, Higgins GA, Bonham T, Tafatia C, et al. Alterations in the human proteome following administration of valproic acid. J Trauma Acute Care Surg. 2016;81:1020–1027. doi: 10.1097/TA.0000000000001249. [DOI] [PubMed] [Google Scholar]
- 157.Yarmohamadi A, Asadi J, Gharaei R, Mir M, Khoshnazar AK. Valproic acid, a histone deacetylase inhibitor, enhances radiosensitivity in breast cancer cell line. J Radiat Cancer Res. 2018;9:86–92. [Google Scholar]
- 158.Soria-Castro R, Schcolnik-Cabrera A, Rodríguez-López G, Campillo-Navarro M, Puebla-Osorio N, Estrada-Parra S et al (2019) Exploring the drug repurposing versatility of valproic acid as a multifunctional regulator of innate and adaptive immune cells. J Immunol Res 2019, Article ID 9678098, 1-24. 10.1155/2019/9678098 [DOI] [PMC free article] [PubMed]
- 159.Kim T, Song S, Park Y, Kang S, Seo H. HDAC inhibition by valproic acid induces neuroprotection and improvement of PD-like behaviors in LRRK2 R1441G transgenic mice. Exp Neurobiol. 2019;28:504–515. doi: 10.5607/en.2019.28.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhu W-W, Lu M, Wang X-Y, Zhou X, Gao C, Qin L-X. The fuel and engine: the roles of reprogrammed metabolism in metastasis of primary liver cancer. Genes Dis. 2020;7:299–307. doi: 10.1016/j.gendis.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li J, Wang T, Xia J, Yao W, Huang F. Enzymatic and nonenzymatic protein acetylations control glycolysis process in liver diseases. FASEB J. 2019;33:11640–11654. doi: 10.1096/fj.201901175R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hosp F, Lassowskat I, Santoro V, De Vleesschauwer D, Fliegner D, Redestig H, et al. Lysine acetylation in mitochondria: from inventory to function. Mitochondrion. 2017;33:58–71. doi: 10.1016/j.mito.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 163.Saunier E, Benelli C, Bortoli S. The pyruvate dehydrogenase complex in cancer: an old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int J Cancer. 2016;138:809–817. doi: 10.1002/ijc.29564. [DOI] [PubMed] [Google Scholar]
- 164.McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, Halim ND, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Woolbright BL, Rajendran G, Harris RA, Taylor JA. Metabolic flexibility in cancer: targeting the pyruvate dehydrogenase kinase:pyruvate dehydrogenase axis. Mol Cancer Ther. 2019;18:1673–1681. doi: 10.1158/1535-7163.MCT-19-0079. [DOI] [PubMed] [Google Scholar]
- 166.Chen J, Guccini I, Di MD, Brina D, Revandkar A, Sarti M, et al. Compartmentalized activities of the pyruvate dehydrogenase complex sustain lipogenesis in prostate cancer. Nat Genet. 2018;50:219–228. doi: 10.1038/s41588-017-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yoneyama K, Shibata R, Igarashi A, Kojima S, Kodani Y, Nagata K, Kurose K, Kawase R, Takeshita T, Hattori S. Proteomic identification of dihydrolipoamide dehydrogenase as a target of autoantibodies in patients with endometrial cancer. Anticancer Res. 2014;34(9):5021–5027. [PubMed] [Google Scholar]
- 168.Brassier A, Ottolenghi C, Boutron A, Bertrand AM, Valmary-Degano S, Cervoni JP, Chrétien D, Arnoux JB, Hubert L, Rabier D, Lacaille F, Keyzer Y, Di Martino V, Lonlay P. Dihydrolipoamide dehydrogenase deficiency: a still overlooked cause of recurrent acute liver failure and Reye-like syndrome. Mol Gen Metab. 2013;109:28–32. doi: 10.1016/j.ymgme.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 169.Neveu J, Hoebeke C, Lebigot E, Naïmi M. Recurrent liver failure in an 11-year-old boy. Clin Chem. 2020;66(8):1115–1123. doi: 10.1093/clinchem/hvaa136. [DOI] [PubMed] [Google Scholar]
- 170.Fei Y, Shi R, Song Z, Wu J. Metabolic control of epilepsy: a promising therapeutic target for epilepsy. Front Neurol. 2020;11:592514. doi: 10.3389/fneur.2020.592514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zsurka G, Kunz WS. Mitochondrial dysfunction and seizures: the neuronal energy crisis. Lancet Neurol. 2015;14(9):956–966. doi: 10.1016/S1474-4422(15)00148-9. [DOI] [PubMed] [Google Scholar]
- 172.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16(1):19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.