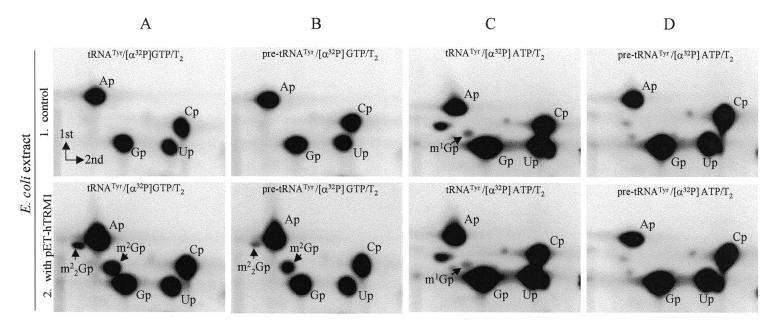
Figure 3.
Autoradiograms of 2-dimensional TLC showing the formation of modified nucleotides m22G and m2G in human tRNATyr and pre-tRNATyr molecules with recombinant hTRM1 enzyme. Unmodified [α-32P]GTP-labeled or [α-32P]ATP-labeled in vitro transcripts of human tRNATyr and pre-tRNATyr were incubated with methyl donor and cell extracts containing or lacking the hTRM1 gene product. The full-length tRNA was isolated, degraded with RNase T2, analyzed by 2-dimensional TLC and spots were identified (see Materials and Methods). The [α-32P] label in the tRNA transcripts and the nuclease used are indicated at the top of each panel. Spots corresponding to m2G and m22G are indicated. (A) [α-32P]GTP-labeled tRNATyr incubated with extract from E.coli pET15b without (A1) and with (A2) the hTRM1 gene. (B) [α-32P]GTP-labeled pre-tRNATyr incubated with extract from E.coli pET15b without (B1) and with (B2) the hTRM1 gene. (C) [α-32P]ATP-labeled tRNATyr incubated with extract from E.coli pET15b without (C1) and with (C2) the hTRM1 gene. (D) [α-32P]ATP-labeled pre-tRNATyr incubated with extract from E.coli pET15b without (D1) and with (D2) the hTRM1 gene. Note that the m1G spots in (C1) and (C2) originated from position G37 (label came from the neighboring A38) and were modified by an inherent E.coli enzyme. In unspliced tRNA G37 is adjacent to the intron and not to a labeled A and therefore no m1G was found in (D1) and (D2).