Abstract
This study investigated the regulation of GRP78 in tumour-associated macrophage polarization in lung cancer. First, our results showed that GRP78 was upregulated in macrophages during M2 polarization and in a conditioned medium derived from lung cancer cells. Next, we found that knocking down GRP78 in macrophages promoted M1 differentiation and suppressed M2 polarization via the Janus kinase/signal transducer and activator of transcription signalling. Moreover, conditioned medium from GRP78- or insulin-like growth factor 1-knockdown macrophages attenuated the survival, proliferation, and migration of lung cancer cells, while conditioned medium from GRP78-overexpressing macrophages had the opposite effects. Additionally, GRP78 knockdown reduced both the secretion of insulin-like growth factor 1 and the phosphorylation of the insulin-like growth factor 1 receptor. Interestingly, insulin-like growth factor 1 neutralization downregulated GRP78 and suppressed GRP78 overexpression-induced M2 polarization. Mechanistically, insulin-like growth factor 1 treatment induced the translocation of GRP78 to the plasma membrane and promoted its association with the insulin-like growth factor 1 receptor. Finally, IGF-1 blockade and knockdown as well as GRP78 knockdown in macrophages inhibited M2 macrophage-induced survival, proliferation, and migration of lung cancer cells both in vitro and in vivo.
Keywords: NSCLC, TAM, JAK1/2, STAT3/6
Introduction
Non-small-cell lung cancer (NSCLC) is one of the most malignant tumours worldwide and has a high incidence, accounting for approximately 85% of all lung cancers, and it is one of the leading causes of cancer-related death [1]. During the initiation, progression and metastasis of NSCLC, the tumour microenvironment (TME) plays an important role [2, 3]. The TME is a complex cellular environment around tumour cells that includes immune cells, fibroblasts, blood vessels, signalling molecules and the extracellular matrix. Some immune cells can be used as predictors of survival, prognosis and recurrence of NSCLC [4]. Recent studies indicate that targeted therapies against these immunological biomarkers in the TME are a novel and appealing approach to treating NSCLC [5–7].
Tumour-associated macrophages (TAMs) are highly abundant in most solid tumour microenvironments and are implicated in all stages of tumour progression [8, 9]. In NSCLC, TAMs actively promote tumorigenesis, growth and metastasis via multiple mechanisms, such as promoting cancer cell initiation and metastasis with various cytokines and chemokines, suppressing the T-cell-mediated immune response and facilitating angiogenesis [10]. However, TAMs are not a single uniform population and are composed of distinct pro- and antitumour subpopulations. Generally, monocytes/macrophages undergo specific differentiation into M1 or M2 macrophages depending on the local environment. M1-polarized macrophages (classically activated macrophages) are induced by interferon-γ (IFN-γ), lipopolysaccharides (LPS), and granulocyte–macrophage colony stimulating factor (GM-CSF) and secrete proinflammatory and immunostimulatory cytokines, including interleukin-1β (IL-1β), tumour necrosis factor-α (TNF-α), IL-12 and IL-23, which contribute to eliciting antigen-specific T helper cell 1 (Th1) and Th17 cell inflammatory responses; M2-polarized macrophages (alternatively activated macrophages) are activated by IL-4, IL-10, and IL-13, produce high levels of IL-10 and tumour growth factor-α (TGF-α) and help to maintain the immunosuppressive TME. After several decades of investigation, it is accepted that TAMs more closely resemble M2-polarized macrophages [11]. Due to their close relationship with malignant tumours, TAMs are now considered potential biomarkers for the diagnosis and prognosis of various cancers. Moreover, TAMs themselves or the polarization pathway are considered potential therapeutic targets for cancer treatments [12, 13].
The 78 kilodalton glucose regulated protein (GRP78), also known as binding-immunoglobulin protein (BiP), is a member of the heat shock protein 70 (HSP70) family of molecular chaperones that have multiple functions [14]. GRP78 is constitutively expressed in the endoplasmic reticulum (ER) lumen and modulates the unfolded protein response (UPR). Moreover, GRP78 functions as a central regulator of ER stress and controls the activation of transmembrane ER stress sensors. Another interesting fact revealed by Misra demonstrates that GRP78 can translocate to cell surface and functions as one co-receptor for low density lipoprotein receptor-related protein (LRP) in α2 macroglobulin-induced signal transduction in macrophages [15]. However, accumulating evidence shows that GRP78 contributes to tumour progression by modulating multiple signalling pathways [16–19], and targeting GRP78 induces apoptosis of some tumour cells [20, 21]. Notably, one early study found that cigarette smoke extract increased the expression of GRP78 in lung cancer cells via the p38/mitogen-activated protein kinase (MAPK) pathway [22]. In NSCLC, GRP78 was upregulated in tumour cells induced by cancer-associated fibroblasts, which then promoted tumour invasion and metastasis [23]. Another report using peripheral blood samples from NSCLC patients demonstrated that GRP78 was highly enriched in late-stage lung cancer and that the overall survival of patients with high GRP78 expression was significantly reduced [24]. Recently, GRP78 was reported to induce M1 polarization in breast cancer [25]. Another recent study revealed that conditioned medium from M2 macrophages increased GRP78 expression in tumour cells and facilitated tumour cell migration [26]. However, the underlying mechanism in NSCLC is still elusive. Thus, we aimed to investigate whether GRP78 exerted the same function on M2 polarization in NSCLC and whether this regulation promotes the progression of NSCLC via enhanced proliferation and migration.
Insulin-like growth factor 1 (IGF-1) is a peptide hormone that mediates cell proliferation, differentiation, and survival after engagement with its receptor, which is expressed by all cells. Substantial evidence demonstrates that IGF-1/IGF-1R signalling is closely related to tumour development and progression [27, 28]. Although some studies have discussed the function of IGF-1R in macrophages [29, 30], the specific effect of IGF-1R on the modulation of M2 polarization is not clear. Moreover, several lines of evidence have shown cross-regulation between IGF-1R and GRP78 [31, 32], and one recent study reported that GRP78 expression was upregulated in pancreatic cancer cells upon treatment with the small molecule ONC-212, while the combination of ONC-212 and the IGF-1R inhibitor AG1024 showed a synergistic effect in suppressing pancreatic cancer cells [33]. Thus, whether IGF-1R and GRP78 coregulate M2 macrophage polarization became a central question in this study.
In the present study, we focused on the function of GRP78 in regulating M2 polarization of macrophages and NSCLC progression. Our study, for the first time, revealed that GRP78 was essential for M2 differentiation, and mechanistic data demonstrated that this effect was dependent on the activation of the JAK/STAT pathway in macrophages. Furthermore, the GRP78 expression level in macrophages influenced the proliferation and migration of cocultured lung cancer cells, possibly through association with IGF-1R in macrophages and regulating the downstream JAK/STAT pathway. IGF-1 neutralization with blocking antibodies not only repressed M2 macrophage polarization but also impaired M2 macrophage-induced proliferation and migration of lung cancer cells. Our findings uncovered a novel function of GRP78 in the regulation of macrophage polarization, and the mechanism revealed may be useful for developing novel therapies to treat NSCLC and other solid cancers by modulating TAM polarization and function.
Materials and methods
Reagents
RPMI 1640 medium, F12K medium and DMEM were purchased from Thermo Fisher Scientific. Lipofectamine 3000, the TRIzol Plus RNA purification kit, SuperScript IV reverse transcriptase, Pierce ECL western blotting substrate, and the Pierce cell surface protein biotinylation and isolation kit were obtained from Thermo Fisher Scientific. Alexa Fluor 488-conjugated mouse anti-human GRP78 antibody, Alexa Fluor 555-conjugated Wheat Germ Agglutinin was purchased from Thermo Fisher Scientific. Alexa Fluor 555-conjugated rabbit anti-human IGF-1R (Tyr1346) antibody was purchased from GmbH online. Rabbit anti-human GRP78 antibody and rabbit anti-human PCNA antibody were purchased from Abcam. Rabbit anti-human STAT3 antibody, rabbit anti-human phospho-STAT3 (Tyr705) antibody, rabbit anti-human STAT6 antibody, rabbit anti-human phospho-STAT6 (Tyr641) antibody, rabbit anti-human JAK1 (6G4) antibody, rabbit anti-human phospho-JAK1 (Tyr1034/1035) antibody, rabbit anti-human JAK2 (D2E12) XP antibody, rabbit anti-human phospho-JAK2 (Tyr1007/1008) (C80C3) antibody, mouse anti-β actin (8H10D10) antibody, rabbit anti-human IGF-1 receptor antibody, and rabbit anti-human phospho-IGF-1 receptor (Tyr1135/1136) (19H7) antibody were obtained from Cell Signalling Technology. Normal rabbit IgG, normal goat anti-mouse IgG-HRP and goat anti-rabbit IgG-HRP antibodies were obtained from Santa Cruz. Human TruStain FcX Fc receptor blocking solution, FITC anti-human F4/80+ antibody, PE anti-human CD86+ antibody, APC anti-human CD11b antibody, PE anti-human CD206 MMR antibody, APC anti-human CD68 antibody, and PE anti-human CD163 antibody were purchased from Biolegend. All primers were synthesized by Shanghai Sangon Biotech (China). LightCycler 480 SYBR Green I Master was obtained from Roche. Ruxolitinib and AG490 were obtained from Selleck. Goat anti-human IGF-1 neutralizing antibody (Cat No: AF-291-NA), normal goat IgG control, the IGF-1 ELISA kit, the IFN-γ ELISA kit, the IL-4 ELISA kit, the IL-13 ELISA kit, GM-CSF, M-CSF, LPS, IFN-γ, IL-4, and IL-13 were purchased from R&D Systems. Phorbol 12-myristate 13-acetate (PMA), 4% polyformaldehyde (PFA), and bovine serum albumin (BSA) were obtained from Sigma-Aldrich. The MTT kit and BCA kit were purchased from Shanghai Sangon Biotech (China). pLKO.1, pLenti-CMV-MCS-GFP-SV-puro, psPAX2 and pMD2. G plasmids were obtained from Addgene. pLP1, pLP2 and pLP/VSVG plasmids were purchased from Thermo Fisher Scientific.
Cell culture
The human acute monocytic leukaemia cell line THP-1 and the human nonsmall cell lung cancer cell lines A549, HCC827 and NCI-H1299 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). THP-1, HCC827 and NCI-H1299 cells were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. A549 cells were maintained in complete F12K medium containing 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin.
Macrophage differentiation, polarization and treatment
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors and isolated by Ficoll density centrifugation. Written consent was obtained from all donors, and the study was designed in accordance with the Declaration of Helsinki and approved by the ethics committee of Central South University in China. To induce macrophage (Mφ) differentiation, CD14+ monocytes were isolated from PBMCs by a classical monocyte isolation kit (Miltenyi Biotech, Germany) and then differentiated into Mφs by coculturing with 25 ng/mL GM-CSF (R&D Systems, USA) for 7 days in complete RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 100 μg/mL streptomycin, and 100 U/mL penicillin. The initial cell density was 6.6 × 105 cells/mL, and the culture medium was replaced every 2 days. The cell viability and density were monitored by ViCell every 2 days before medium replacement. THP-1 cells were first differentiated into Mφs by incubation with 50 ng/mL PMA for 24 h. After differentiation, the expression of panmacrophage markers (CD11b, F4/80) was assessed by FACS. Macrophages with viability higher than 90% were used for gene knockdown or overexpression by lentivirus at day 8 and were then used to perform macrophage polarization at day 10.
PBMC-derived Mφs or THP-1-derived Mφs were polarized to M1 or M2 macrophages by exposing the cells to 100 ng/mL LPS plus 20 ng/mL IFN-γ or 20 ng/mL IL-4 plus 20 ng/mL IL-13 for 48 h, respectively. Cell viability and density were measured by ViCell after 48 h of polarization. Medium replacement was also performed with complete RPMI 1640 medium after viability and density measurements. Then, the expression of M1 macrophage markers (CD86) and M2 macrophage markers (CD163, CD206) was assessed by FACS.
To assess the effect of conditioned medium (CM) from lung cancer cells on M2 macrophage polarization, CM derived from A549, HCC827 and NCI-H1299 cells was collected after 72 h of culture and then mixed with fresh RPMI 1640 medium containing 10% FBS (for HCC827 and NCI-H1299 cells) or F12K medium supplemented with 10% FBS (for A549 cells) at a ratio of 1:2 (CM/fresh medium). Nutrient-sufficient CM was then used to treat PBMC- or THP-1 cell-derived Mφs for 48 h. To block IGF-1 signalling, IGF-1 neutralizing antibodies were added to the culture medium at a concentration of 5 μg/mL during the M2 polarization process.
Plasmid construction and transfection
Human GRP78 and IGF-1 genes were cloned from HeLa cDNAs with the primers listed in Table 1. Full-length GRP78 was then subcloned into multiple cloning sites of pLenti6.3/V5-DEST expression plasmids, which were transfected into 293 T cells using Lipofectamine 3000, together with pLP1 and pLP2 packaging plasmids and pLP/VSVG envelope plasmid, to produce GRP78-expressing lentivirus. The lentivirus titre was determined by HIV-p24-based ELISA (ab218268, Abcam, UK) and was adjusted to 1 × 108 TU/mL, which was utilized to infect macrophages at an MOI = 100 to generate GRP78-overexpressing cells. The expression of GRP78 in macrophages was assessed by intracellular staining for GRP78 and FACS analysis.
Table 1.
Forward and reverse primers used in plasmid construction
| Primer name | Sequence (5ʹ–3ʹ) |
|---|---|
| GRP78 forward | AAAAAAATGAAGCTCTCCCTGGTGGCCGC |
| GRP78 reverse | AAAAAACTACAACTCATCTTTTTCTGCTGTATCCTCTTC |
| IGF-I forward | AAAAAAATGGGAAAAATCAGCAGTCTTC |
| IGF-I reverse | AAAAAATTACTTGCGTTCTTCAAATGTACTTC |
ShRNAs targeting GRP78 and IGF-1 and negative control shRNA were synthesized by Shanghai GenePharma and subcloned into the pLKO.1-EGFP-Puro vector. The target sequences of the shRNAs are listed in Table 2. The pLKO.1-shGRP78-EGFP-Puro and pLKO.1-shIGF-1-EGFP-Puro plasmids were then transfected into HEK293T cells with psPAX2 packaging plasmid and pMD2. G envelope plasmid using Lipofectamine 3000 to produce shRNA-containing lentivirus. The lentivirus titre was determined by HIV-p24-based ELISA (ab218268, Abcam, UK) and was then adjusted to 1 × 108 TU/mL, which was used to infect PBMC-derived or THP-1-derived macrophages at an MOI = 100 to obtain GRP78- or IGF-1-knockdown cells. The transfection efficiency of shRNAs was analysed by measuring the expression of EGFP with FACS, and the knockdown efficiency was assessed by qPCR and Western blotting.
Table 2.
Target sequence of GRP78 shRNA
| ShRNA No | Target sequence |
|---|---|
| shRNA#1 | gctcgactcgaattccaaaga |
| shRNA#2 | gccaccaagatgctgacattg |
| shRNA#3 | gaaatcgaaaggatggttaat |
| shRNA#4 | ctgggtacatttgatctgact |
Flow cytometry analysis of macrophage polarization
CD14+ monocyte- or THP-1-derived macrophages were harvested, washed with PBS and incubated with Human TruStain FcX Fc receptor blocking reagent for 10 min at room temperature. The cells were stained with FITC anti-human F4/80, APC anti-human CD11b and PE anti-human CD86 antibodies or FITC anti-human F4/80, APC anti-human CD11b and PE anti-human CD206 antibodies at 4 °C for 1 h in the dark. Then, the cells were rinsed with PBS twice and fixed with 4% PFA for 10 min at 4 °C. The expression levels of F4/80, CD11b and CD86 or F4/80, CD11b and CD206 were measured using a FACS LSRII (BD Biosciences), and the results were analysed with FlowJo V10 software. The gating strategy for M1 and M2 macrophages was as follows: (1) F4/80+CD11b+ population for macrophages; (2) CD11b+CD86+ population for M1 macrophages and CD11b+CD206+ population for M2 macrophages.
The experimental control information was listed as follows: Fig. 2A, B: Control cells were CD14+ monocyte- or THP-1-derived macrophages without any treatments; Fig. 3E: PBMC-derived macrophages cultured in normal medium without IL4 + IL13; Fig. 3J: PBMC-derived macrophages transduced with empty vector and cultured in normal medium without IL4 + IL13; Fig. 4C: PBMC-derived macrophages transduced with empty vector and cultured in normal medium without IL4 + IL13; Fig. 6F: PBMC-derived macrophages transduced with empty vector and cultured in normal medium without IL4 + IL13 or antibody; Fig. 9E: PBMC-derived macrophages transduced with shNC or shIGF1 and cultured in normal medium without IL4 + IL13. All the above control cells were stained with the same antibodies as the experimental cells.
Fig. 2.
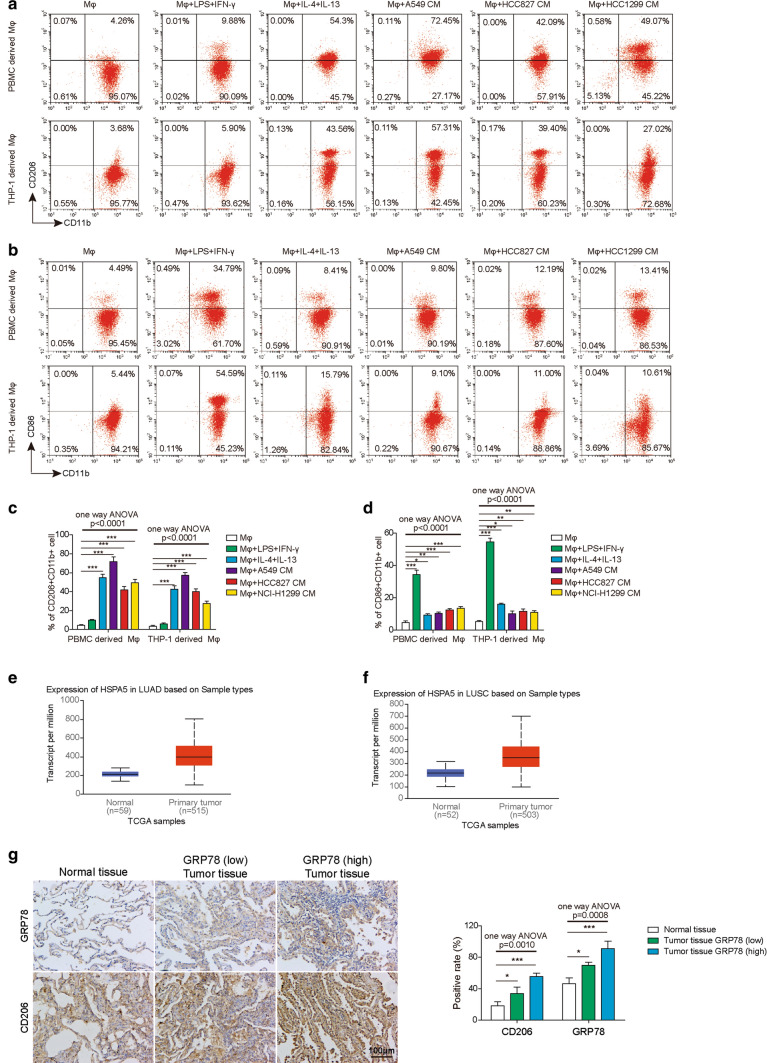
Upregulation of GRP78 in macrophages upon M2 polarization. A Representative flow cytometry image of M2 macrophages in each group is shown. B Representative flow cytometry image of M1 macrophages in each group is shown in each group. C The proportions of M2 macrophages in each group were determined by flow cytometry. N = 6. D The proportions of M1 macrophages in each group were determined by flow cytometry. N = 6. E The expression of GRP78 in human lung adenocarcinoma (LUAD) tumour samples and normal tissues in TCGA database. F The expression of GRP78 in human lung squamous cell carcinoma (LUSC) tumour samples and normal tissues in TCGA database. G The expression of GRP78 and CD206 in human lung tumour samples and normal tissues by IHC staining. All results are representative of three independent experiments. Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test (C, D). ***p < 0.001, **p < 0.01, *p < 0.05
Fig. 3.
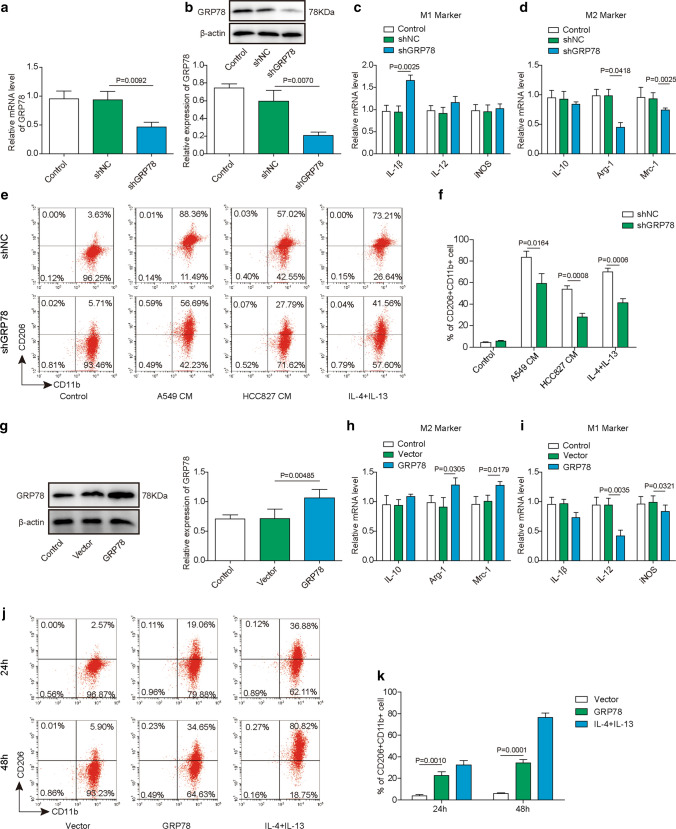
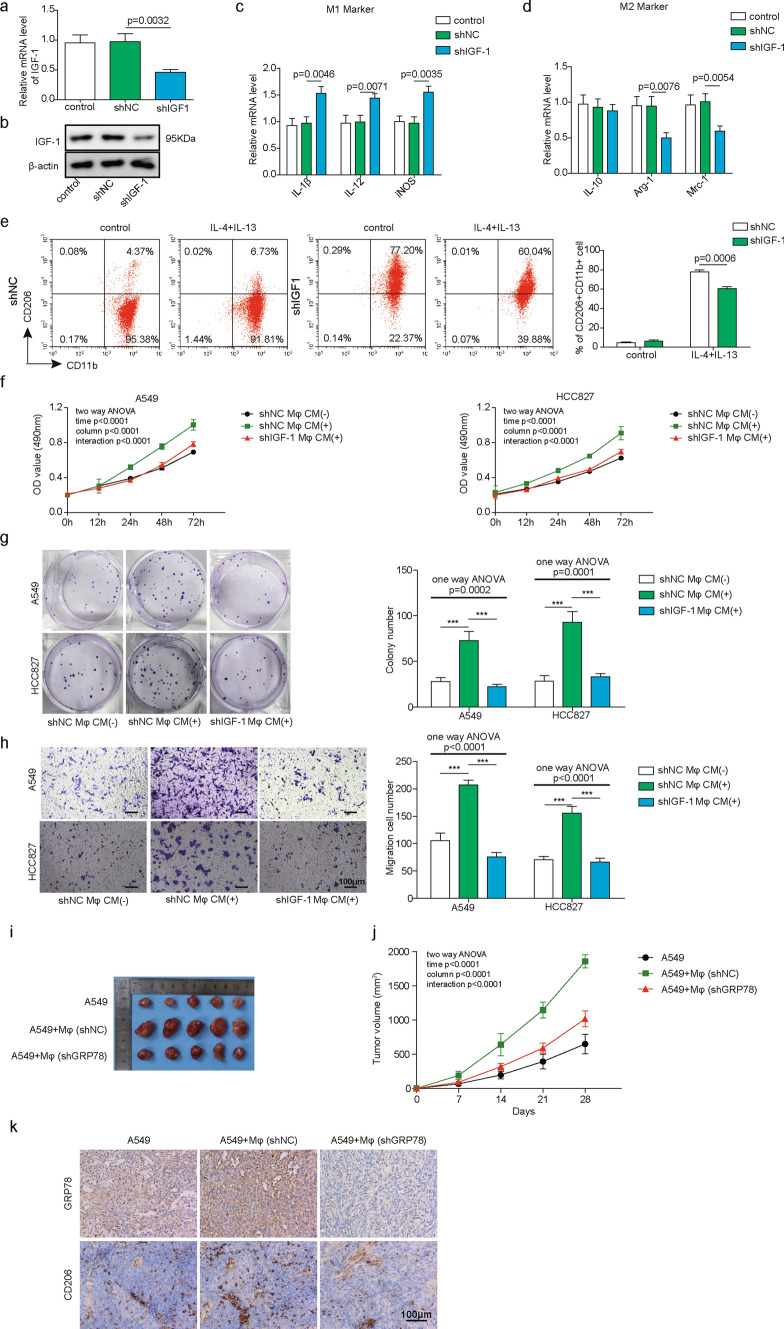
GRP78 is required for the M2 polarization of macrophages. A–F PBMC-derived macrophages were transfected with the indicated shRNAs by lentivirus. A The relative expression of GRP78 in each group of cells was measured by qPCR. N = 6. B The knockdown efficiency of GRP78 was further verified by western blotting, and β-actin was used as the loading control in western blotting. N = 6. C, D After shRNA transduction, PBMC-derived macrophages were polarized to M1 or M2 macrophages for 48 h. The relative expression of the M1 markers IL-1β, IL-12 and iNOS (C) and the M2 markers IL-10, Arg-1 and Mrc-1 (D) was determined by qPCR. N = 6. E, F shRNA-transfected cells were then cocultured with control medium, A549 CM, HCC827 CM or IL-4/IL-13 for another 48 h. E The polarization of M2 macrophages was analysed by FACS using the M2 surface marker CD11b+CD206+. N = 6. F Statistical analysis of the proportions of M2 macrophages in E. G–K Macrophages were untransfected or transfected with the indicated plasmid. G The expression of GRP78 was assessed by western blotting, and β-actin was used as the loading control. N = 6. The normalized GRP78 expression to β-actin is shown on the right. H–I After transfection, PBMC-derived macrophages were polarized to M1 or M2 macrophages for 48 h. The relative expression of the M2 markers IL-10, Arg-1, and Mrc-1 (H) and the M1 markers IL-1β, IL-12, and iNOS (I) was determined by qPCR. N = 6. J, K After transfection, PBMC-derived macrophages were then cultured without or with IL-4 and IL-13 for 48 h. J The polarization of M2 macrophages was analysed by FACS using the M2 surface marker CD11b+CD206+. N = 6. K Statistical analysis of the proportions of M2 macrophages in J. A, C, D, H, I mRNA expression was normalized to that of GAPDH, and the experiments were performed in triplicate. A–J The results are representative of three independent experiments. A, C, D, F, H, I, K Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test (A, C, D, H, I, K) or unpaired t test (F). ***p < 0.001, **p < 0.01, *p < 0.05
Fig. 4.
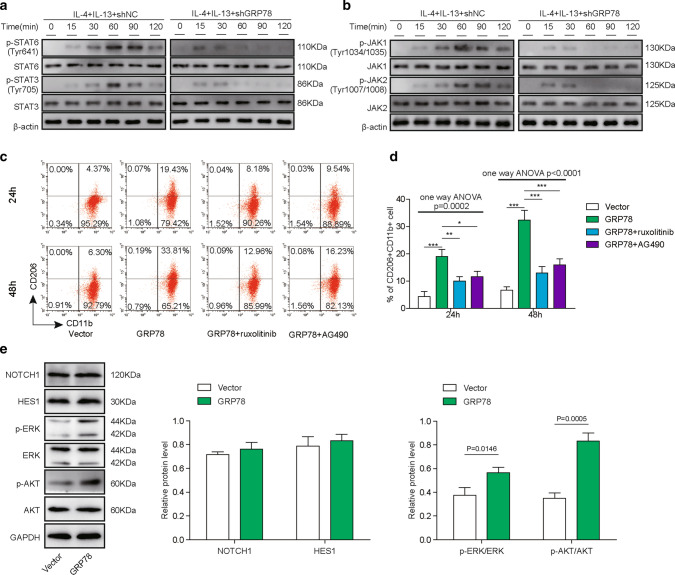
GRP78-induced M2 macrophage polarization is dependent on JAK/STAT activation. A, B PBMC-derived macrophages were transfected with the indicated shRNAs and then cultured for the indicated time in the presence of 20 ng/mL IL-4 and 20 ng/mL IL-13. The phosphorylation and total content of STAT6 and STAT3 (A) and JAK1 and JAK2 (B) were measured by western blotting. β-actin was used as the loading control. N = 6. C, D Macrophages were transfected with the indicated plasmid and then cultured for 24 h or 48 h without or with the JAK1/2 inhibitor ruxolitinib at 100 nM or the JAK1 inhibitor AG490 at 10 μM. C The polarization of M2 macrophages was analysed by FACS. N = 6. D Statistical analysis of the proportions of M2 macrophages in C. E PBMC-derived macrophages were transfected with the empty vector or GRP78-overexpressing plasmid and then cultured in the presence of 20 ng/mL IL-4 and 20 ng/mL IL-13 for 48 h. The expression of NOTCH1 and HES1 and the phosphorylation and total content of AKT and ERK were measured by western blotting, and GAPDH was used as the loading control. N = 6 A–E The results are representative of three independent experiments. D, E Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. ***p < 0.001, **p < 0.01, *p < 0.05
Fig. 6.
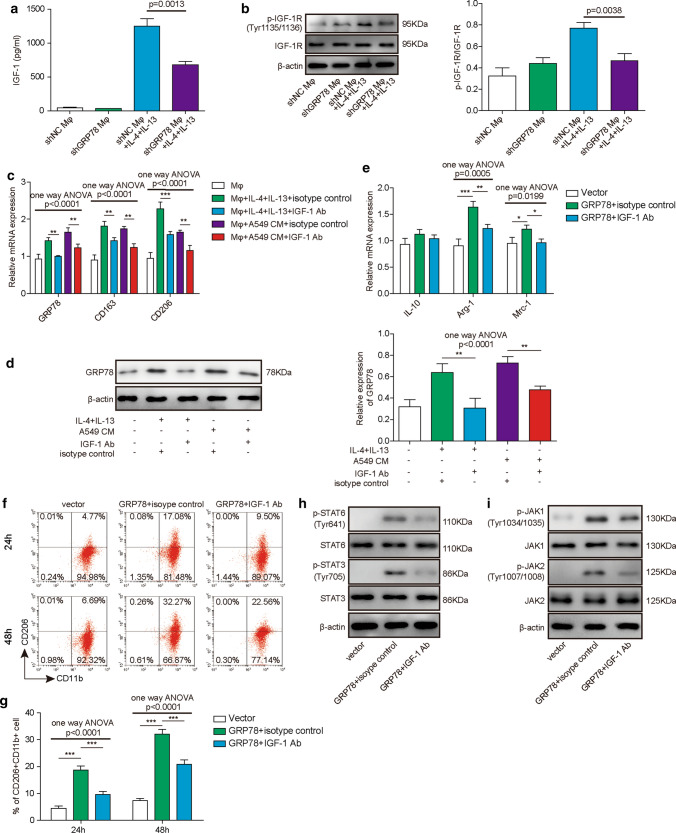
The IGF-1/IGF-1R signaling pathway is implicated in GRP78-induced M2 macrophages. A, B PBMC-derived macrophages were transfected with the indicated shRNAs and then cultured in the absence or presence of IL-4/IL-13 for 48 h. A The concentration of secreted IGF-1 in the supernatant was assessed by ELISA. N = 6. B The phosphorylation and total content of IGF-1R were measured by western blotting, and β-actin was used as the loading control. N = 6. C, D Macrophages were cultured alone or with IL-4/IL-13 or A549 CM in the presence of the IGF-1 neutralizing antibody or isotype control antibody for 48 h. C The relative expression of GRP78, CD163 and CD206 was determined by qPCR. The mRNA expression was normalized to that of GAPDH, and the experiments were performed in triplicate. N = 6. D The expression of GRP78 protein was assessed by western blotting, and β-actin was used as the loading control. N = 6. E–I Macrophages were transfected with the indicated plasmid and then cultured with isotype control antibody or IGF-1 neutralizing antibody for 24 h or 48 h, respectively. E The relative expression of IL-10, Arg-1 and Mrc-1 was measured by qPCR after 48 h of culture. The mRNA expression was normalized to that of GAPDH, and the experiments were performed in triplicate. N = 6. F The polarization of M2 macrophages was analysed by FACS after 24 h or 48 h of culture. N = 6. G Statistical analysis of M2 polarization in F. H–I The phosphorylation and total content of STAT6 and STAT3 (H) and JAK1 and JAK2 (I) were measured by western blotting. β-actin was used as the loading control. (A–F, H–I) The results are representative of three independent experiments. (A, C, E, G) Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. ***p < 0.001, **p < 0.01, *p < 0.05
Fig. 9.
IGF-1 is required for polarization and the proliferation and migration of lung cancer cells. A–H PBMC-derived macrophages were transfected with the indicated plasmid by lentivirus. A The relative expression of IGF-1 in macrophages was measured by qPCR. The mRNA expression was normalized to that of GAPDH, and the experiments were performed in triplicate. N = 5. B The knockdown efficiency of IGF-1 was further verified by western blotting, and β-actin was used as the loading control. N = 5. C–E After shRNA transduction, PBMC-derived macrophages were polarized to M1 or M2 macrophages for 48 h. C, D The relative expression of the M1 markers IL-1β, IL-12 and iNOS (C) and the M2 markers IL-10, Arg-1 and Mrc-1 (D) was determined by qPCR. The mRNA expression was normalized to that of GAPDH, and the experiments were performed in triplicate. N = 5. E After shRNA transduction, PBMC-derived macrophages were polarized to M2 macrophages for 48 h, which was then analysed by FACS and statistical analysis of the proportions of M2 macrophages. N = 5. F–H Forty-eight hours later, macrophage CM was collected to culture A549 and HCC827 cells for the indicated assays. F The proliferation of A549 and HCC827 cells was assessed by MTT assay at the indicated times. N = 5. G The survival and proliferation of A549 and HCC827 cells were determined by colony formation assay and statistical analysis of the colony numbers. N = 5. H The migration of A549 and HCC827 cells was measured by Transwell assay (left panel). Scale bar, 50 μm. The statistical analysis of the migrated cells is shown in the right panel. I–K Six-week-old nude mice were subcutaneously injected with 2 × 106 A549 cells, a mixture of 2 × 106 A549 cells plus 1 × 106 shNC-transfected M2 macrophages, or a mixture of 2 × 106 A549 cells and 1 × 106 shGRP78-transfected M2 macrophages. N = 5. I Four weeks later, the mice were euthanized, and the tumour tissues were dissected for analysis. J The tumour growth kinetics were monitored by measuring the tumour volume every 7 days. K The expression of GRP78 in xenografted NSCLC tumour samples and in tumour-associated macrophages was measured by IHC. Scale bar, 100 μm. A–E, G–H The results are representative of three independent experiments. F, I, J The results are representative of five independent experiments. A, C, D, F, G, H, J, K, M Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) (A, C, D, G, H), two-way ANOVA (F, J) or by unpaired two-tailed t test (E). ***p < 0.001, **p < 0.01, *p < 0.05
RNA extraction and quantitative real-time PCR
Total RNA was isolated from macrophages that were lysed and homogenized in TRIzol reagent. cDNA was obtained via reverse transcription PCR with oligo dT(20) primers. qPCR was performed using a LightCycler 480 SYBR Green I Master and LightCycler LC480 real-time qPCR instrument (Roche). The relative mRNA expression levels of Grp78, Cd86, Cd163, Il-1b, Il-10, Il-12, Inos, Arg-1 and Mrc-1 were normalized to that of Gapdh. All experiments were performed in triplicate. The primers used for qPCR are listed in Table 3.
Table 3.
The primers used in quantitative real-time PCR
| Primer name | Sequence (5ʹ–3ʹ) |
|---|---|
| Grp78 forward | CATCACGCCGTCCTATGTCG |
| Grp78 reverse | CGTCAAAGACCGTGTTCTCG |
| Cd68 forward | TGGGGCAGAGCTTCAGTTG |
| Cd68 reverse | TGGGGCAGGAGAAACTTTGC |
| Cd163 forward | TTTGTCAACTTGAGTCCCTTCAC |
| Cd163 reverse | TCCCGCTACACTTGTTTTCAC |
| Il-1b forward | ATGATGGCTTATTACAGTGGCAA |
| Il-1b reverse | GTCGGAGATTCGTAGCTGGA |
| Il-10 forward | GACTTTAAGGGTTACCTGGGTTG |
| Il-10 reverse | TCACATGCGCCTTGATGTCTG |
| Il-12 forward | CCTTGCACTTCTGAAGAGATTGA |
| Il-12 reverse | ACAGGGCCATCATAAAAGAGGT |
| Inos forward | AGGGACAAGCCTACCCCTC |
| Inos reverse | CTCATCTCCCGTCAGTTGGT |
| Arg-1 forward | TGGACAGACTAGGAATTGGCA |
| Arg-1 reverse | CCAGTCCGTCAACATCAAAACT |
| Mrc-1 forward | GGGTTGCTATCACTCTCTATGC |
| Mrc-1 reverse | TTTCTTGTCTGTTGCCGTAGTT |
| Gapdh forward | CTGACTTCAACAGCGACACC |
| Gapdh reverse | GTGGTCCAGGGGTCTTACTC |
Membrane protein extraction by biotinylation for western blotting
Macrophages were first rinsed twice with cold PBS and then incubated with 1.0 mg/mL EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, diluted in PBS) for 30 min at 4 °C on a shaker. The cells were washed three times with 100 mM glycine in PBS to remove excess biotin reagent and byproducts. Then, the cells were lysed in RIPA buffer supplemented with proteinase and phosphatase inhibitors. After centrifugation at 12,000 rpm at 4 °C for 10 min, the clarified cell lysates were subsequently used to pulldown biotin-labelled membrane proteins via incubation with 50 μL of streptavidin-conjugated sepharose beads at 4 °C for 4 h on a rotator. The precipitated sepharose beads were washed three times with lysis buffer, and the biotin-labelled membrane proteins were released from the beads by heating with SDS loading buffer for 10 min. The protein samples were then used to perform western blotting.
Western blotting
The protein content in the cell lysate was quantified using a BCA kit. Samples containing 50 μg of protein were boiled by mixing with 4 × reducing loading buffer for 10 min, resolved by SDS-PAGE and transferred to active PVDF membranes (Millipore Corp, Bedford, MA), which were then blocked with 5% BSA in Tris-buffered saline with 0.1% Tween 20 (TBST) for 1 h. The PVDF membranes were then rinsed and incubated with the following primary antibodies at 4 °C overnight: anti-GRP78 (1:1000), anti-PCNA (1:1000), anti-β actin (1:10,000), anti-STAT3 (1:2000), anti-p-STAT3 (Tyr705) (1:1000), anti-STAT6 (1:2000), anti-p-STAT6 (Tyr641) (1:1000), anti-JAK1 (1:2000), anti-p-JAK1 (1:1000), anti-JAK2 (1:2000), anti-p-JAK2 (1:1000), anti-IGF-1 (1:2000), and anti-p-IGF-1 (Tyr35/T36) (111:1000). After washing, the PVDF membranes were incubated with goat anti-mouse IgG-HRP (1:5000) or goat anti-rabbit IgG-HRP (1:5000) secondary antibodies for 1 h. After washing with TBST, the blots were detected with ECL substrate and imaged by an iBright FL1000 imaging system (Thermo Fisher Scientific).
Cell surface protein biotinylation and co-immunoprecipitation assay
Macrophages were first biotinylated using the Pierce cell surface protein biotinylation and isolation kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions, rinsed with cold PBS, collected, and lysed in 1 × cell lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 × proteinase inhibitor cocktail) by rotation at 4 °C for 1 h. After centrifugation at 12,000 g and 4 °C for 10 min, the cleared supernatants were collected in a new 1.5 mL tube. Next, the supernatant was transferred to a NeutrAvidin Agarose column and incubated by end-over-end mixing on a rotator for 30 min at room temperature. After washing four times, the biotinylated proteins on the resin were eluted by elution buffer containing 10 mM DTT. The eluted proteins were then subjected to a co-immunoprecipitation assay.
Cell surface protein samples or total protein samples were aliquoted into four tubes, which were then incubated with anti-GRP78, anti-IGF-1R or normal rabbit IgG by gentle rotation at 4 °C for 4 h. Next, 40 μL of prewashed Protein A/G-conjugated agarose beads was added to capture the precipitated antibody-antigen complexes via 2 h incubation at 4 °C. After centrifugation and washing the beads three times, the protein samples were eluted with 1 × reducing loading buffer and subjected to western blotting analysis.
MTT assay
A549 and HCC827 cells were seeded in 96-well plates at a density of 104 cells per well in 100 µL of macrophage CM supplemented with 5 μg/mL isotype control or 5 μg/mL IGF-1 neutralizing antibody, and one plate of cells without macrophage CM was cultured at the same time as a reference. For each treatment, five replicate wells and one blank control group with culture medium only were prepared. The cells were then incubated at 37 °C in a humidified incubator with 5% CO2. Cell viability was monitored by MTT assay for 3 days following the manufacturer’s instructions. Three independent experiments were performed, and one representative result is shown in the figures.
Cell colony formation assay with soft agar
A 6-well plate was precoated with a layer of solidified medium containing 0.8% agarose, and then 1 × 104 A549 or HCC827 cells were plated on this layer in 2.5 mL of macrophage-derived CM supplemented with 0.4% agarose. After culturing for 14 days, cell colonies were fixed and stained with 0.1% crystal violet solution. After washing with PBS, the whole well was captured, and the number of viable colonies larger than 0.1 mm was calculated with ImageJ software (NIH, USA).
Cell migration assay
Transwell assays were performed to assess cell migration. Briefly, 5 × 104 A549 or HCC827 cells in 500 μL macrophage CM were seeded in Transwell inserts (Corning), and the lower chambers were filled with 500 μL of complete F12K (A549) or RPMI 1640 (HCC827) medium as a chemoattractant. After incubation for 15 h, cells that migrated to the lower chamber were fixed with 4% PFA and stained with 0.1% crystal violet. Stained cells were then imaged on an IX71 inverted microscope (Olympus, Japan), and six random fields per well were captured. The cell number per well was calculated in the captured images using ImageJ software (NIH, USA).
Enzyme-linked immunosorbent assay (ELISA)
Control, GRP78-knockdown or GRP78-overexpressing macrophages were seeded into 6-well plates (5 × 104 per well) in 2.5 mL of fresh complete RPMI 1640 medium. To induce M2 differentiation, 20 ng/mL IL-4 and 20 ng/mL IL-13 were loaded as indicated. Then, these cells were cultured for 48 h. The concentrations of IGF-1, IFN-γ, IL-4 and IL-13 in the supernatants were measured with the human IGF-1 Quantikine ELISA Kit (R&D), human IFN-γ Quantikine ELISA Kit (R&D), human IL-4 Quantikine ELISA Kit (R&D) and human IL-13 Quantikine ELISA Kit (R&D), respectively. The ELISA experimental procedure followed the manufacturer’s instructions.
Xenograft tumour model in nude mice
Nude mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. and were maintained in pathogen-free facilities at Central South University. Mice of matched age and sex were used in all experiments, and they were randomly allocated to experimental groups. All protocols for animal experiments were approved by the Animal Care and Use Committee (ACUC) of Central South University. The maximal tumour volumes were in accordance with the guidelines of ACUC.
To establish a tumour growth model, shNC- or shGRP78-transfected M2 macrophages were trypsinized, washed with PBS and filtered through a 40-μm cell strainer. Then, 2 × 106 A549 cells, a mixture of 2 × 106 A549 cells plus 1 × 106 shNC-transfected M2 macrophages (THP-1-derived macrophages that were then polarized into M2 macrophages by IL4 and IL13), or a mixture of 2 × 106 A549 cells and 1 × 106 shGRP78-transfected M2 macrophages were transplanted into 6-week-old nude mice via subcutaneous injection. The tumour growth kinetics were monitored by measuring the tumour volume every 7 days with a Vernier calliper. Tumour volume was calculated as length × width × height. Mice with tumour sizes larger than 20 mm at the longest axis were euthanized based on ethical considerations. After 4 weeks, the tumour tissues were dissected from euthanized mice, imaged and analysed. This dissection was designed in accordance with the Declaration of Helsinki and approved by the ethics committee of Central South University in China.
Immunofluorescent staining
After IGF-1 treatment, macrophages cultured on poly-l-lysine-coated cover glasses were washed with PBS once and then fixed with 4% paraformaldehyde for 15 min at room temperature. After washing, the cells were permeabilized with 0.1% Triton X-100 for 15 min at room temperature. Next, the cells were blocked with 2% BSA for 30 min and then stained with Alexa Fluor 488-conjugated mouse anti-human GRP78 antibody (1:25, Thermo Fisher Scientific, MA, USA) and Alexa Fluor 555-conjugated rabbit anti-human IGF-1R (Tyr1346) antibody (1:50, antibodies-online GmbH, Aachen, Germany) or Alexa Fluor 555-conjugated Wheat Germ Agglutinin (WGA, which was used as the plasma membrane marker. 1:50, Thermo Fisher Scientific, MA, USA) at 4 °C for 6 h. To display the cell nuclei, 100 μg/mL DAPI was applied to incubate the cells for 15 min at room temperature. After washing, the cells were subjected to an inverted fluorescence microscope (Olympus IX73, Tokyo, Japan) for imaging. Five individual fields were chosen for imaging.
Immunohistochemistry (IHC)
Human lung cancer tissues and peritumour tissues were first cut into small pieces, fixed and cryoprotected in 30% sucrose for 24 h. Frozen samples were then sectioned into pieces at 6 μm thickness. Sectioned specimens were attached onto slides, washed and incubated with 3% H2O2 in methanol for 15 min to quench endogenous peroxidase. After washing, the sections were blocked and probed with the indicated primary antibodies at 4 °C overnight, including rabbit anti-human CD206 and rabbit anti-human GRP78 antibodies. After washing, the sections were probed with goat anti-rabbit IgG-HRP for 1 h at room temperature, followed by incubation with DAB substrate for signal development. Finally, these sections were washed, dehydrated, sealed with coverslips and imaged by inverted fluorescence microscopy (IX-53, Olympus). Generally, approximately five randomly chosen fields were captured for each sample. Data were collected from five independent experiments.
Human lung cancer tissues and peritumour tissues were collected from NSCLC patients during surgery. Written consent was obtained from all donors, and the study was designed in accordance with the Declaration of Helsinki and approved by the ethics committee of Central South University in China.
Statistical analysis
All experiments were performed three times, with one representative experiment shown. The data are represented as the mean ± standard deviation (SD). Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software, Inc.). Unpaired two-tailed Student’s t tests were used to compare the differences between two groups. One-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used for multiple comparisons. Two-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used for comparing the difference of multiple curves. Statistical significance was determined as indicated in the figure legends. *, **, and *** denote significance at the 0.05, 0.01, and 0.001 levels, respectively.
Results
GRP78 is upregulated during M2 macrophage polarization
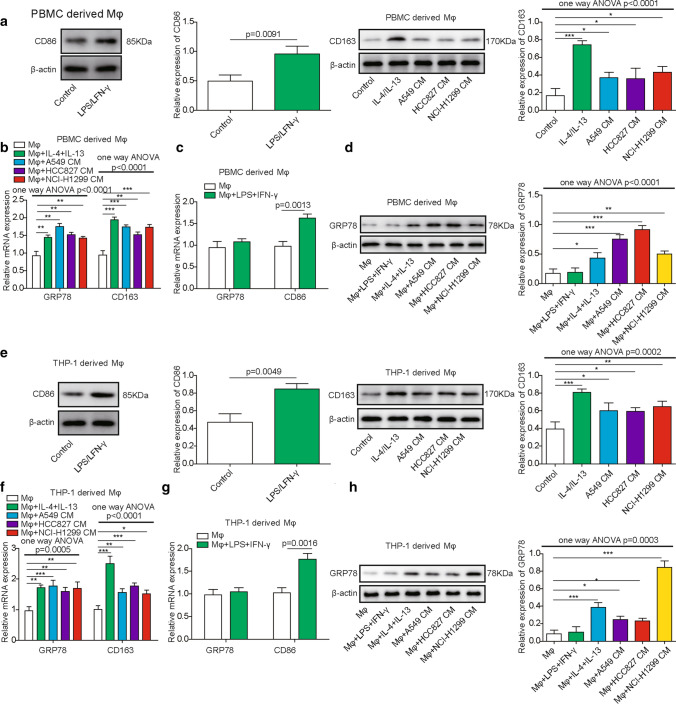
To determine the expression of GRP78 during macrophage differentiation, we first treated PBMC- and THP-1-derived macrophages with LPS/IFN-γ or IL-4/IL-13 to induce M1 or M2 polarization. To determine the effect of TME on macrophage differentiation, we also cultured PBMC- and THP-1-derived macrophages in CM of A549, HCC827, and NCI-H1299 lung cancer cells. Afterwards, polarized macrophages were collected for western blotting to analyse the expression of CD86 and CD163, which are markers of M1 and M2 macrophages, respectively. From Fig. 1A, E, we can see that LPS/IFN-γ treatment induced the upregulation of CD86 in PBMCs or THP-1-derived macrophages, whereas IL-4/IL-13 treatment resulted in the significant elevation of CD163. Interestingly, CM from A549, HCC827 and NCI-H1299 cells also facilitated the polarization of M2 macrophages in our experiments, which was supported by the milder augmentation of CD163 when compared with that in control cells (Fig. 1A, E). Consistently, qPCR results also showed that CD163 was significantly upregulated in polarized macrophages upon IL-4/IL-13 treatment or coculture with CM from A549, HCC827 and NCI-H1299 cells (Fig. 1B, F). In addition, CD86 expression was elevated in polarized macrophages after the administration of LPS/IFN-γ (Fig. 1C, G). GRP78 expression was increased in M2 macrophages and was not affected during M1 macrophage differentiation (Fig. 1B, C, F, G). Moreover, we performed western blotting to measure the protein level of GRP78 during M2 macrophage polarization or upon tumour CM treatment, and the results showed that GRP78 was indeed upregulated at the translational level, which was consistent with the qPCR results (Fig. 1D, H). Moreover, we analyzed the proportion of M1 macrophages and M2 macrophages by flow cytometry. The results showed that the M2 (CD206 + CD11b +) proportion was significantly upregulated by IL-4/IL-13 and CM from A549, HCC827 and NCI-H1299 cells (Fig. 2A, C), while the M1 (CD86 + CD11b +) proportion was significantly augmented by LPS/IFN-γ and slightly induced by CM from A549, HCC827 and NCI-H1299 cells (Fig. 2B, D). Overall, these data revealed that GRP78 was upregulated in macrophages upon M2 polarization in both cytokine-induced and tumour cell-induced conditions. Additionally, we confirmed that GRP78 was upregulated in the LUAD and LUSC databases from TCGA database (Fig. 2E, F). To explore the physiological relevance of our study, we then collected several human lung cancer samples and performed IHC experiments to measure the expression of GRP78 and to assess the level of the M2 macrophage marker CD206. The data demonstrated that when the GRP78 protein expression level was upregulated, the level of CD206 in M2 macrophages was enhanced. Moreover, the higher the level of GRP78 was, the higher the expression of CD206, which indicates that GRP78 facilitated M2 macrophages in human lung cancer tissues (Fig. 2G).
Fig. 1.
Upregulation of GRP78 in macrophages upon M2 polarization. A–D Human PBMC-derived macrophages were generated from CD14+ monocytes and then polarized to M1 or M2 macrophages for 48 h. In parallel, tumour cell-derived CM was used to culture macrophages, which were pretreated with GM-CSF or M-CSF, for 48 h. A After M1 or M2 polarization, the expression of CD86 or CD163 in M1 or M2 macrophages was assessed by western blotting. β-actin was used as the loading control. N = 6. B The relative expression of GRP78 and CD163 in PBMC-derived macrophages in response to the indicated treatments was assessed by qPCR. N = 6. C The relative expression of GRP78 and CD86 in macrophages treated without or with LPS/IFN-γ was measured by qPCR. N = 6. D The expression of GRP78 in PBMC-derived macrophages in response to the indicated treatments was determined by western blotting, and β-actin was used as the loading control. N = 6. E–H THP-1 cells were first differentiated into Mφs for 24 h and were then polarized to M1 or M2 macrophages for 48 h. E The expression of CD86 or CD163 in M1 or M2 macrophages was assessed by western blotting. β-actin was used as the loading control. N = 6. The normalized expression of CD86 or CD163 to β-actin in macrophages in response to the indicated treatments is shown on the right in histograms. F Tumour cell-derived CM was used to culture macrophages, which were pretreated with GM-CSF or M-CSF, for 48 h. The relative expression of GRP78 and CD163 in THP-1-derived macrophages after the indicated treatments was assessed by qPCR. N = 6. G The relative expression of GRP78 and CD86 in THP-1-derived macrophages treated without or with LPS/IFN-γ was measured by qPCR. N = 6. H The expression of GRP78 in THP-1-derived macrophages in response to the indicated treatments was determined by western blotting, and β-actin was used as the loading control. N = 6. The normalized expression of GRP78 to β-actin in macrophages in response to the indicated treatments is shown in the right histograms. All results are representative of three independent experiments. Error bars represent the mean ± SD. p values were determined by unpaired two-tailed t test (A, C, E, G) or one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test (A, B, D, E, F, H). ***p < 0.001, **p < 0.01, *p < 0.05
GRP78 is required for M2 macrophage polarization
To investigate whether GRP78 is essential for M2 polarization of macrophages, we knocked down GRP78 in PBMC-derived macrophages using shRNAs. Both qPCR and western blotting results showed that shRNA knockdown led to the significant downregulation of GRP78 at the transcriptional and translational levels compared with that in negative control shRNA-transfected cells (Fig. 3A, B). Then, we analysed the relative expression of M1 marker genes, including IL-1β, IL-12, and iNOS, and M2 marker genes, including IL-10, Arg-1, and Mrc-1, using qPCR. The data demonstrated that the expression of the M1 marker gene IL-1β was obviously increased, and IL-12 was slightly upregulated, while iNOS expression was not affected. The increased IL-1β indicated that GRP78 silencing led to enhanced M1 macrophage polarization. In contrast, the M2 marker genes Mrc-1 and Arg-1 were repressed after silencing GRP78, while IL-10 expression was not affected (Fig. 3C, D). Next, we cultured shNC- or shGRP78-transfected macrophages in culture medium (CM) from A549 and HCC827 cells and CM in supplemented with IL-4/IL-13, and we analysed the M2 polarization of macrophages by flow cytometry. Consistently, CM from both A549 and HCC827 cells induced potent M2 polarization of macrophages, as indicated by high proportions of CD11b+CD206+ cells when compared with that in medium supplemented with IL-4/IL-13. However, GRP78 knockdown evidently suppressed the polarization of M2 macrophages compared with that in shNC-transfected cells (Fig. 3E, F). To further verify that GRP78 is essential for M2 macrophage differentiation, we overexpressed GRP78 to determine whether it could promote this process. The GRP78-overexpressing plasmid was transfected into macrophages via lentivirus, and western blot analysis confirmed the ectopic expression of GRP78 (Fig. 3G). As expected, GRP78 overexpression resulted in robust upregulation of the M2 macrophage marker genes Arg-1 and Mrc-1 as well as a reduction in the M1 macrophage marker genes IL-1β and iNOS (Fig. 3H, I). Moreover, GRP78 overexpression significantly enhanced the differentiation of macrophages into the M2 subset compared with that of control cells (Fig. 3J–K). Overall, our data revealed that M2 polarization of macrophages was impaired upon GRP78 silencing but enhanced by GRP78 overexpression, which indicates that GRP78 is required for this process.
GRP78-induced M2 polarization of macrophages is dependent on JAK/STAT pathway activation
A previous study found that the JAK-STAT pathway is activated during macrophage polarization [34], and different cytokines or stimuli induce distinct JAK/STAT signalling pathways in the macrophage reprogramming process, which finally produces M1 or M2 macrophages [35]. Moreover, accumulating evidence has revealed that GRP78 participates in the modulation of the JAK-STAT pathway [36, 37]. To explore the underlying mechanism of GRP78-induced M2 polarization, we first used IL-4/IL-13 to treat shNC- or shGRP78-transfected PBMC-derived macrophages to induce their polarization and then assessed the activation of the JAK-STAT pathway by western blotting. The results clearly showed that IL-4/IL-13 induced activation of the JAK-STAT pathway, as evidenced by the increased phosphorylation of STAT3, STAT6, JAK1 and JAK2 in shNC-transfected cells; however, the phosphorylation was severely impaired after GRP78 knockdown, indicating suppression of the JAK-STAT pathway (Fig. 4A, B). To further confirm the crucial role of the JAK-STAT pathway in GRP78-induced M2 macrophage polarization, we administered the JAK1/2 inhibitor ruxolitinib or the JAK1 inhibitor AG490 to GRP78-overexpressing macrophages and then used FACS to measure the M2 differentiation of macrophages after 24 h or 48 h, respectively. The results indicated that blocking JAK-STAT pathway activation potently restrained M2 macrophage polarization compared with that of GRP78-overexpressing cells (Fig. 4C, D). In addition, we examined several canonical signalling pathways involved in M2 macrophage polarization, such as the NOTCH1 pathway, AKT pathway, and ERK pathway, in response to GRP78 overexpression in macrophages. The Western blotting results showed that the expression of NOTCH1 and downstream HES1 underwent no obvious changes, whereas the phosphorylation of AKT and ERK was significantly enhanced (Fig. 4E), which suggested that GRP78 might regulate the polarization of M2 macrophages through the AKT and ERK pathways; however, this mechanism was not the focus of the present study. In summary, our data suggest that activation of the JAK-STAT pathway is indispensable for GRP78-induced M2 polarization of macrophages.
GRP78 expression in macrophages affects the proliferation and migration of lung cancer cells
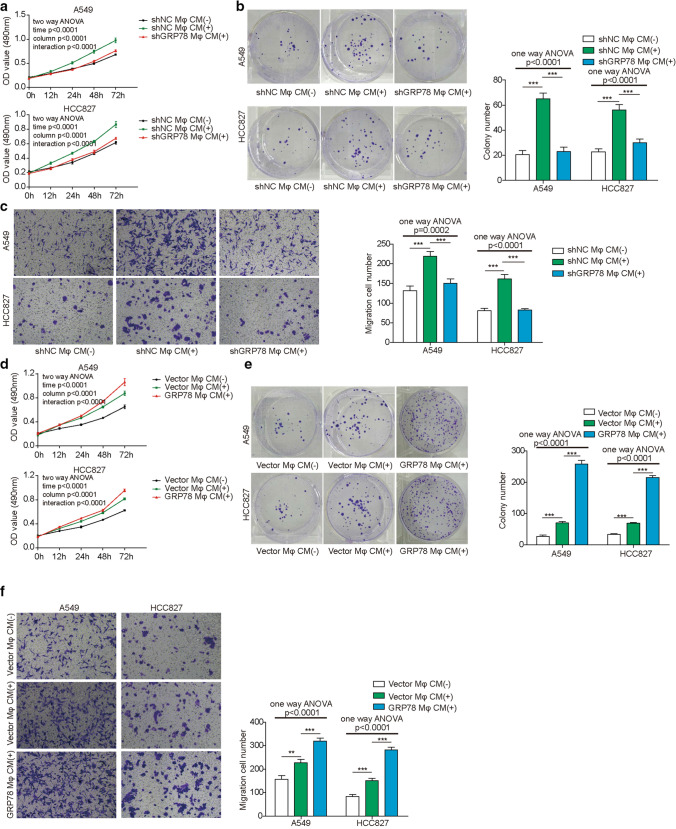
TAMs have been found to play multiple roles during lung cancer progression and metastasis [10]. To verify the function of GRP78 in TAMs and its effect on lung cancer cells, we cultured A549 or HCC827 lung cancer cells in CM from shNC- or shGRP78-transfected macrophages. First, cell viability was determined by MTT assay, and the data showed that CM from shNC-transfected macrophages promoted the survival of both A549 and HCC827 cells; however, GRP78 knockdown abrogated the effect of macrophage CM on lung cancer cell survival (Fig. 5A). Next, we performed a colony formation assay to assess the proliferation capacity of A549 and HCC827 cells in different CMs. Consistently, these two types of cells exhibited similar proliferation patterns in macrophage-derived CMs, which were enhanced in CM from shNC-transfected macrophages but dampened in CM from GRP78-silenced macrophages (Fig. 5B). Furthermore, we measured cell migration capacity using A549 and HCC827 cells cultured in macrophage-derived CM, and the results illustrated that GRP78 overexpression in macrophages augmented the migration of A549 and HCC827 cells; in contrast, CM from GRP78-silenced macrophages completely abolished this effect (Fig. 5C). In addition, we overexpressed GRP78 in macrophages, obtained CM and implemented similar experiments as described above. As expected, ectopic expression of GRP78 in macrophages led to a significant increase in the survival, proliferation, and migration of A549 and HCC827 cells compared with those of control cells (Fig. 5D–F).
Fig. 5.
GRP78 expression in macrophages affects the proliferation and migration of lung cancer cells. PBMC-derived macrophages were transfected with the indicated shRNAs (A–C) or plasmids (D–F), and then they were polarized to M2 macrophages for 48 h. CM was collected to culture A549 and HCC827 cells with fresh RPMI 1640 medium at a ratio of 1:1 for the indicated assays. A, D The proliferation of A549 and HCC827 cells was assessed by MTT assay at the indicated times. N = 5. B, E The survival and proliferation of A549 and HCC827 cells were determined by colony formation assay and statistical analysis of the colony numbers. N = 5. C, F The migration of A549 and HCC827 cells was measured by Transwell assay and statistical analysis of the migrated cells. N = 5. The results are representative of three independent experiments. Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) (B, C, E, F) or two-way ANOVA (A, D) followed by Tukey’s post hoc test. ***p < 0.001, **p < 0.01, *p < 0.05
The IGF-1/IGF-1R signalling pathway is involved in GRP78-induced M2 polarization of macrophages
Previous studies have reported that IGF-1/IGF-1R signalling is involved in macrophage differentiation [29, 38]; moreover, IGF-1/IGF-1R is closely associated with GRP78 expression. For example, Yin et al. reported that cell surface GRP78 facilitated hepatoma cell proliferation and migration by activating IGF-1R, while IGF-1 could induce the expression of GRP78 via the PI3K and MAPK signalling pathways [31]. This positive feedback loop might have a crucial function in macrophage polarization. To ascertain the possible association between IGF-1/IGF-1R signalling and GRP78 during M2 macrophage polarization, we cultured control or GRP78-knockdown macrophages in vitro in the absence or presence of IL-4/IL-13 and measured the IGF-1 concentration in the supernatant by ELISA. To our surprise, the data showed that IL-4/IL-13-induced M2 polarization dramatically increased the secretion of IGF-1 by macrophages; however, GRP78 knockdown partially reduced its expression (Fig. 6A). Next, we determined the activation status of the IGF-1 pathway in macrophages by examining the phosphorylation of IGF-1R, and western blot results showed that IGF-1R phosphorylation was augmented in shNC-transduced macrophages after IL-4 plus IL-13 treatment, however, additional GRP78 knockdown partially impaired the increase of IGF-1R phosphorylation (Fig. 6B). Based on these results, we next investigated whether IGF-1 is required for M2 polarization of macrophages. As shown in Fig. 6C, IL-4/IL-13 or A549 CM treatment induced the upregulation of GRP78, CD163 and CD206; however, the addition of IGF-1 neutralizing antibody robustly attenuated the expression of these factors, as measured by qPCR (Fig. 6C). Moreover, the IL-4/IL-13 or A549 CM treatment-induced upregulation of GRP78 protein expression was also suppressed by an IGF-1 neutralizing antibody in western blot experiments (Fig. 6D). We aimed to explore the effect of IGF-1 blockade on GRP78-induced M2 polarization by culturing GRP78-overexpressing macrophages with isotype control antibody or IGF-1 neutralizing antibody. qPCR experiments demonstrated that the relative expression of M2 subset marker genes, including Arg-1 and Mrc-1, was downregulated by IGF-1 neutralization (Fig. 6E). Consistently, IGF-1 deprivation reduced M2 macrophage polarization by almost 30–40% compared with that in the isotype control antibody-treated group (Fig. 6F, G). In addition, IGF-1 antibody treatment attenuated the GRP78 overexpression-induced phosphorylation of STAT3, STAT6, JAK1, and JAK2 (Fig. 6H, I). Overall, these data revealed that the IGF-1/IGF-1R pathway was activated during GRP78-induced M2 polarization, while blocking this pathway partially suppressed GRP78-induced polarization.
GRP78 is recruited to the plasma membrane and interacts with IGF-1R upon IGF-1 treatment
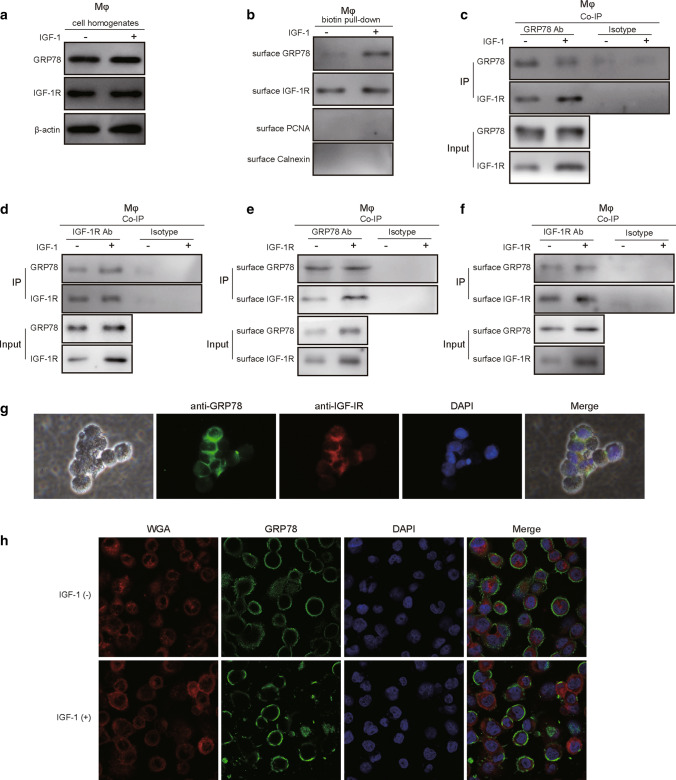
To investigate the detailed mechanism by which IGF-1/IGF-1R regulates GRP78-induced M2 polarization, we first treated macrophages with or without IGF-1 for 24 h, and western blot experiments using cell lysates showed that IGF-1 administration had no obvious influence on the expression of GRP78 or IGF-1R (Fig. 7A). We next performed biotinylation and streptavidin pulldown of cell surface proteins, and the results demonstrated that IGF-1 exposure induced the plasma membrane translocation of GRP78, while the cell surface level of IGF-1R had no significant change (Fig. 7B). Moreover, the Co-IP experiment using total cell lysate and the GRP78 antibody showed that much more IGF-1R interacted with GRP78 in the presence of IGF-1 than in the absence of IGF-1 (Fig. 7C). Consistently, reverse Co-IP with total cell lysate and the IGF-1R antibody demonstrated that more GRP78 interacted with IGF-1R in response to IGF-1 stimulation (Fig. 7D). To further demonstrate the interaction between GRP78 and IGF-1, we then used biotinylated cell membrane proteins to conduct Co-IP experiments with GRP78 antibody or IGF-1R antibody. Subsequent western blotting data showed that GRP78 interacted with the intracellular domain of IGF-1R upon IGF-1 stimulation (Fig. 7E, F). Finally, we performed immunofluorescence staining of GRP78 and IGF-1R in IGF-1-treated macrophages, and the results showed that IGF-1 treatment induced significant membrane translocation of GRP78 and clear colocalization of GRP78 and IGF-1R (Fig. 7G). Furthermore, we compared the translocation of GRP78 in macrophages in the absence or presence of IGF-1, the immunofluorescence data elucidated that IGF-1 treatment indeed induced the translocation of GRP78 to plasma membrane (Fig. 7H). In summary, these experiments demonstrated that cytosolic GRP78 was recruited to the plasma membrane and interacted with IGF-1R in macrophages after IGF-1 stimulation.
Fig. 7.
IGF-1 induces the translocation of GRP78 to the plasma membrane and its association with IGF-1R. A, B PBMC-derived macrophages were treated with or without 100 ng/mL IGF-1 for 24 h, and then the cell surface proteins were isolated via biotinylation and streptavidin pulldown. A Total cell lysates were analysed by western blotting to determine the expression of GRP78 and IGF-1R, and β-actin was used as the loading control. N = 4. B Pulled-down cell surface proteins were subjected to western blotting to assess the amount of GRP78 on the plasma membrane. The nuclear marker protein PCNA and ER marker protein calnexin were utilized as indicators to show the purity of the surface protein fractions. N = 4. C, D Macrophages were treated with or without 10 ng/mL IGF-1 for 24 h, and then the cells were lysed and subjected to immunoprecipitation using antibodies against GRP78 (C) or IGF-1R (D). The associated IGF-1R (C) or GRP78 (D) was analysed by western blotting. N = 4. (E–F) Macrophages were treated without or with IGF-1 for 24 h, and then the cell surface proteins were isolated via biotinylation and streptavidin pulldown. Cell surface proteins were then used to perform immunoprecipitation with antibodies against GRP78 (E) or IGF-1R (F), and the associated IGF-1R (E) or GRP78 (F) was analysed by western blotting. N = 4. G Macrophages were fixed, permeabilized and stained with Alexa Fluor 488-conjugated mouse anti-human GRP78 antibody plus Alexa Fluor 555-conjugated rabbit anti-human IGF-1R antibody, which were then imaged with fluorescence microscopy. Cell nuclei were stained with DAPI. Scale bar, 20 μm. H Macrophages were treated without or with IGF-1 for 24 h, then they were fixed, permeabilized and stained with Alexa Fluor 488-conjugated mouse anti-human GRP78 antibody plus Alexa Fluor 555-conjugated Wheat Germ Agglutinin (WGA, which was used as the plasma membrane marker), which were then imaged with fluorescence microscopy. Cell nuclei were stained with DAPI. Scale bar, 20 μm. A–H The results are representative of three independent experiments
The proliferation and migration of lung cancer cells is repressed by IGF-1 neutralization
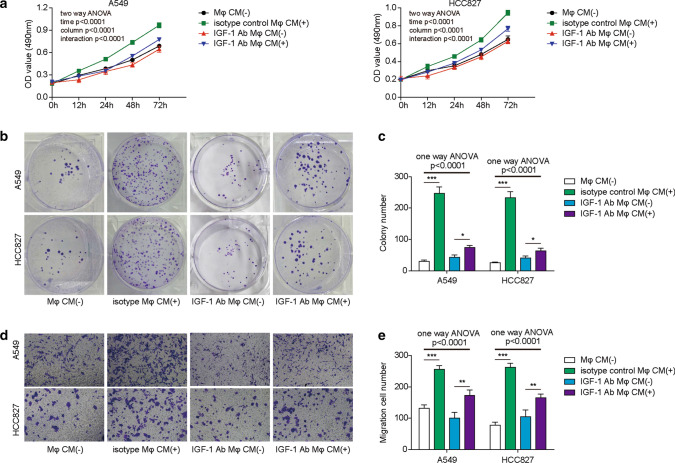
Accumulating evidence has proven that M2-like TAMs promote the proliferation and migration of lung cancer cells [39, 40]. Since GRP78 is required and essential for M2 macrophage polarization, we then wanted to explore the effect of blocking IGF-1 on the proliferation and migration of lung cancer cells, which is closely related to tumour progression and metastasis. First, A549 or HCC827 lung cancer cells were cultured with normal medium or macrophage-derived CM in the absence or presence of the IGF-1 neutralizing antibody. Subsequent MTT assays demonstrated that both A549 and HCC827 cells showed accelerated proliferation in macrophage-derived CM compared with that of normal medium; however, the addition of the IGF-1 neutralizing antibody impaired the proliferation of both types of cells (Fig. 8A). Consistently, the survival and proliferation of A549 and HCC827 cells in the colony formation assay also showed a similar pattern; there were many more colonies in macrophage-derived CM than in normal medium, while IGF-1 blockade potently repressed cell proliferation and reduced colony numbers (Fig. 8B, C). Finally, we utilized a Transwell assay to address whether tumour cell migration was also affected by IGF-1 blockade. As expected, the data illustrated that macrophage-derived CM robustly enhanced the migration of A549 and HCC827 cells when compared with that of normal medium, and the addition of the IGF-1 neutralizing antibody significantly abrogated the migratory capacities of A549 and HCC827 cells (Fig. 8D, E). Taken together, our data revealed that IGF-1 neutralization suppressed the M2 macrophage-promoted proliferation and migration of lung cancer cells.
Fig. 8.
IGF-1 blockade in M2 macrophages suppresses the proliferation and migration of lung cancer cells. A–E After induction, M2 macrophages were cultured in RPMI 1640 medium for 48 h. Then, CM was collected for subsequent experiments. A A549 and HCC827 cells were cultured in normal medium, or macrophage CM supplemented with 5 μg/mL isotype control or 5 μg/mL IGF-1 neutralizing antibody. The proliferation of A549 and HCC827 cells was assessed by MTT assay at the indicated times. N = 5. B The survival and proliferation of A549 and HCC827 cells were determined by colony formation assay after 14 days of culture. N = 5. C Statistical analysis of the colony numbers in C. D The migration of A549 and HCC827 cells was measured by Transwell assay after 15 h of culture. N = 5. E Statistical analysis of the migrated cells in E. A, C, E The results are representative of three independent experiments. A, C, E Error bars represent the mean ± SD. p values were determined by one-way analysis of variance (ANOVA) (C, E) or two-way ANOVA (A) followed by Tukey’s post hoc test. ***p < 0.001
Endogenous IGF-1 and GRP78 are required for M2 polarization and the proliferation and migration of lung cancer cells
To further ascertain the origin of IGF-1, we first knocked down IGF-1 in macrophages by shRNA and then assessed macrophage polarization, as well as the proliferation and migration of lung cancer cells (A549 and HCC827 cells), in macrophage-derived conditioned medium. Both the qPCR and western blot results showed that the expression of IGF-1 was significantly reduced after shRNA-mediated knockdown (Fig. 9A, B). Subsequently, we found that knocking down IGF-1 in macrophages induced the upregulation of M1 marker genes and the downregulation of M2 marker genes (Fig. 9C, D). Consistently, IGF-1-knockdown in macrophages significantly reduced M2 polarization upon IL-4/IL-13 induction compared with that of control macrophages, which was confirmed by FACS analysis (Fig. 9E). Next, we collected CM from macrophages with or without IGF-1-knockdown and then cultured A549 and HCC827 lung cancer cells. The MTT and colony formation assays showed that a lack of IGF-1 impaired the macrophage-mediated promotion of tumour cell proliferation (Fig. 9F, G). The Transwell assay demonstrated that lung cancer cells exhibited weakened migratory capacity in CM from IGF-1-knockdown macrophages (Fig. 9H). To verify whether the expression of endogenous GRP78 in macrophages affects NSCLC tumour growth in vivo, we then established a xenograft mouse model by subcutaneous injection of A549 cells or A549 cells plus GRP78-sufficient or GRP78-deficient M2 macrophages. Four weeks later, the tumour size data showed that GRP78-sufficient M2 macrophages potently promoted NSCLC tumour growth in vivo, and GRP78-silenced M2 macrophages also mildly enhanced tumour growth compared with the control group (Fig. 9I). Consistently, tumour volume growth kinetics data demonstrated that mice transplanted with A549 and GRP78-sufficient M2 macrophages had the fastest tumour growing speed, whereas mice transplanted with A549 and GRP78-silenced M2 macrophages had a moderate tumour growing speed, and mice receiving only A549 cells showed the slowest tumour growing speed (Fig. 9J). Moreover, we assessed the expression level of GRP78 in xenografted tumour samples and tumour-associated macrophages by IHC, and the data clearly showed that GRP78 was upregulated in xenografted NSCLC tumour samples derived from A549 and GRP78-sufficient M2 macrophages in nude mice. Moreover, the expression overlap between CD206 and GRP78 in IHC further demonstrated that GRP78 expression in TAMs was also elevated in xenografted NSCLC tumours derived from A549 and GRP78-sufficient M2 macrophages (Fig. 9K). Taken together, these experiments illustrated that endogenous IGF-1 and GRP78 in macrophages were indispensable for M2 polarization and the proliferation and migration of lung cancer cells.
Discussion
In the present study, we revealed the novel mechanism by which GRP78 mediates M2 macrophage polarization through IGF-1/IGF-1R signalling. We found that the expression of GRP78 was upregulated during M2 differentiation and was essential for M2 polarization. GRP78 exerted its function by binding to IGF-1R upon IGF-1 stimulation, which subsequently activated the JAK/STAT pathway in macrophages. M2 macrophages facilitated the proliferation and migration of lung cancer cells, which was suppressed by blocking IGF-1 and knockdown experiments.
Previous studies have provided conflicting evidence about the role of TAMs in human lung cancer. Some reports found that there was a positive correlation between TAM infiltration and a good prognosis [41–44]; however, most of these studies confirmed that significantly higher stromal M1 macrophage densities existed in patients with good survival. On the other hand, substantial evidence proved that TAMs were correlated with poor prognosis in lung cancer patients [45–47]. As a chaperone protein in the ER, GRP78 is involved in signal transduction that regulates tumour proliferation, invasion and metastasis [17, 23, 26, 48].
Previous work has demonstrated that UPR-related proteins, including GRP78, PERK, ATF6, and IRE1, are all involved in macrophage polarization. Knockdown of GRP78 or PERK promotes M1 polarization in breast cancer [25], while inhibition of IRE1 prevents ER expansion and restrains M2 polarization [49].
In our study, we found that knockdown of GRP78 led to enhanced IL-1β expression, which indicated that M1 macrophage polarization was augmented. Since a previous study revealed that GRP78 has anti-inflammatory properties and can impair the production of lipopolysaccharide-induced proinflammatory cytokines (including IL-1β) in dendritic cells [50], our study further confirmed the anti-inflammatory function of GRP78 in macrophages. ATF6 is highly expressed in IL-4/IL-13-induced M2 macrophages with the expansion of the ER, while IRE1-XBP inhibition indirectly affects ATF6 cleavage or activation, which also contributes to the suppression of M2 macrophages [49]. However, the underlying mechanism has not been investigated. In the present work, we uncovered that GRP78 promotes the M2 macrophage polarization process, which is consistent with a previous study and reveals an additional mechanism. We confirmed that JAK and STAT proteins are pivotal factors in M1 and M2 macrophage polarization in response to various cytokines [51, 52]. Moreover, GRP78 has been implicated in the regulation of the JAK/STAT pathway. For example, Mina Thon and colleagues reported that GRP78 was required for insulin-enhanced leptin-induced STAT3 signalling [36]. Yun et al. found that GRP78 enhances the phosphorylation of JAK2 and STAT3 to promote the proliferation and migration of mesenchymal stem cells after triggering by crypto [37]. In this study, we identified that cell surface GRP78 modulates the JAK/STAT pathway to potentiate the M2 polarization of macrophages, which is a crucial downstream event after the activation of the IGF-1 receptor. In addition, surface GRP78 blockade by monoclonal antibodies on various tumour cells significantly suppresses PI3K/Akt signalling in tumour cells and impairs the progression and metastasis of tumours [53], which further shows the broad effects of GRP78 on various signalling pathways. Thus, we hypothesized that surface GRP78 neutralization not only attenuates M2 polarization but also directly represses the proliferation and migration of tumour cells, which should have synergetic effects in curing different cancers.
IGF-1R signalling plays an essential role in macrophage polarization. Spadaro and colleagues found that IGF-1R knockout in mice led to elevated adiposity and insulin resistance after administration of a high-fat diet for a certain period; moreover, ablation of IGF-1R reduced the transcription associated with M2-like macrophage activation [29]. Barrett et al. confirmed that IGF-1 was necessary for the full adoption of the M2 phenotype in macrophages, while its blockade impaired the ability of IL-4 to induce activation of AKT to elevate the expression of some M2-associated molecules [38]. Interestingly, GRP78 was reported as a downstream target of IGF-1 signalling, whose expression is regulated by the PI3K/AKT/mTORC1 axis upon IGF-1R activation [32]. Moreover, Yin and colleagues revealed that there was a reciprocal regulation between GRP78 expression and IGF-1R pathway activation; IGF-1 induces GRP78 expression in hepatoma cells, while GRP78 promotes IGF-1R phosphorylation and activation [31]. In the present study, we found that IGF-1 was required for the regulation of macrophage M2 polarization by GRP78. First, the secretion of IGF-1 by macrophages was increased upon M2 differentiation, and the engagement of IGF-1R induced the translocation of GRP78 to the plasma membrane and the association with IGF-1R, as evidenced by the cell imaging experiment and co-IP assay. Second, the interaction between IGF-1R and GRP78 further enhanced the phosphorylation of IGF-1R and activation of downstream pathways, which led to increased M2 hallmark genes, supporting the commitment to M2 polarization. Moreover, knockdown of IGF-1 in macrophages significantly suppressed M2 polarization and impaired M2 macrophage-mediated promotion of the proliferation and migration of NSCLC cells. Consistently, in vivo data further demonstrated that the GRP78 expression level in M2 macrophages could significantly influence NSCLC tumour growth and that normal endogenous GRP78 expression in M2 macrophages could robustly increase the tumour growth speed. The association between the expression level of GRP78 and the infiltration of M2 macrophages in human lung cancer tissue was also confirmed in this study. Overall, our study, for the first time, revealed that IGF-1 was involved in the M2 macrophage-mediated promotion of proliferation and migration of NSCLC cells. In addition to IGF-1, M2 macrophages also secrete vascular endothelial growth factor (VEGF), tumour growth factor-β (TGF-β) and platelet-derived growth factor (PDGF) to promote angiogenesis and metastasis [54, 55]; moreover, M2 macrophages can secrete matrix metalloproteinase-9 (MMP-9) and urokinase-type plasminogen activator to facilitate the invasion and metastasis of tumour cells into surrounding tissues [56]. In terms of immune suppression, IL-10 is the most-studied cytokine produced by M2 macrophages, along with IL-13 and TGF-β, which can repress the antitumour immune response and promote tumour growth [57]. Although M2 macrophage-secreted IGF-1 showed potent effects on tumour cell proliferation and migration, the detailed mechanism underlying this phenotype still merits further investigation.
Although we have uncovered a novel mechanism of GRP78 in M2 polarization, there were still some limitations in this study. On the one hand, many previous reports have focused on the function of GRP78 in tumour cells; however, the effect of GRP78 on tumour cell-induced macrophage polarization has not been sufficiently investigated. On the other hand, previous studies have found that colorectal cancer cells can secrete IGF-1 and express IGF-1 receptors, which stimulate colorectal cancer development and progression via cell autonomous and microenvironmental effects [58]. Therefore, reciprocal regulation between tumour cells and macrophages should exist, although we only focused on the influence of IGF-1 derived from macrophages on tumour cells. The regulation of M2 TAM polarization by IGF-1 produced from tumour cells deserves further investigation. Finally, we explored the function of GRP78 in macrophages via in vitro experiments. However, the TME is an extremely complicated milieu, and whether this effect still occurs in vivo requires more work in the future. In addition, based on previous reports [25], GRP78 might also regulate macrophage polarization via downstream PERK or IRE1 signalling independent of IGF-1/IGF-1R.
In summary, our work revealed that GRP78 was increased and essential for M2 macrophage polarization. GRP78 promotes this differentiation by activating the JAK/STAT pathway in an IGF-1/IGF-1R-dependent manner, which is upregulated in M2 polarization and recruits more GRP78 to the plasma membrane to interact with IGF-1R. The association between GRP78 and IGF-1R further enhances IGF-1R phosphorylation and the downstream JAK/STAT pathway. This positive regulatory loop not only facilitates M2 polarization but also boosts the proliferation and migration of lung cancer cells. However, blocking IGF-1 signalling successfully restrains the proliferation and migration of lung cancer cells promoted by M2 macrophages. Overall, the mechanism revealed here has enormous potential to become a therapeutic target for controlling TAM commitment and tumour progression.
Funding
This work was supported by the National Natural Science Foundation of China (81802290) and National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases (Lung Cancer, grant number: z027002).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The study was designed in accordance with the Declaration of Helsinki and approved by the ethics committee of Central South University in China.
Consent to participate
Written consent was obtained from all donors.
Consent for publication
The informed consent obtained from study participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, et al. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 2.Graves EE, Maity A, Le QT. The tumor microenvironment in non-small-cell lung cancer. Semin Radiat Oncol. 2010;20(3):156–163. doi: 10.1016/j.semradonc.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40(4):558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu K, Okita R, Nakata M. Clinical significance of the tumor microenvironment in non-small cell lung cancer. Ann Transl Med. 2013;1(2):20. doi: 10.3978/j.issn.2305-5839.2013.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuster MM. Targeting the lung cancer microenvironment: harnessing host responses. In: Takiguchi Y, editor. Molecular targeted therapy of lung cancer. Singapore: Springer Singapore; 2017. pp. 309–327. [Google Scholar]
- 6.Mittal V, El Rayes T, Narula N, McGraw TE, Altorki NK, Barcellos-Hoff MH. The microenvironment of lung cancer and therapeutic implications. Adv Exp Med Biol. 2016;890:75–110. doi: 10.1007/978-3-319-24932-2_5. [DOI] [PubMed] [Google Scholar]
- 7.Owusu BY, Thomas S, Venukadasula P, Han Z, Janetka JW, Galemmo RA, Jr, et al. Targeting the tumor-promoting microenvironment in MET-amplified NSCLC cells with a novel inhibitor of pro-HGF activation. Oncotarget. 2017;8(38):63014–63025. doi: 10.18632/oncotarget.18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu SQ, Xu R, Li XF, Zhao XK, Qian BZ. Prognostic roles of tumor associated macrophages in bladder cancer: a system review and meta-analysis. Oncotarget. 2018;9(38):25294–25303. doi: 10.18632/oncotarget.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol. 2017;10(1):58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99(Pt B):180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Cassetta L, Kitamura T. Targeting tumor-associated macrophages as a potential strategy to enhance the response to immune checkpoint inhibitors. Front Cell Dev Biol. 2018;6:38. doi: 10.3389/fcell.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuiderweg ER, Hightower LE, Gestwicki JE. The remarkable multivalency of the Hsp70 chaperones. Cell Stress Chaperones. 2017;22(2):173–189. doi: 10.1007/s12192-017-0776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra UK, Gonzalez-Gronow M, Gawdi G, Hart JP, Johnson CE, Pizzo SV. The role of Grp 78 in alpha 2-macroglobulin-induced signal transduction. Evidence from RNA interference that the low density lipoprotein receptor-related protein is associated with, but not necessary for, GRP 78-mediated signal transduction. J Biol Chem. 2002;277(44):42082–42087. doi: 10.1074/jbc.M206174200. [DOI] [PubMed] [Google Scholar]
- 16.Li R, Yanjiao G, Wubin H, Yue W, Jianhua H, Huachuan Z, et al. Secreted GRP78 activates EGFR-SRC-STAT3 signaling and confers the resistance to sorafeinib in HCC cells. Oncotarget. 2017;8(12):19354–19364. doi: 10.18632/oncotarget.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao X, Liu H, Zhang X, Zhang L, Li X, Wang C, et al. Cell surface GRP78 accelerated breast cancer cell proliferation and migration by activating STAT3. PLos One. 2015;10(5):e0125634. doi: 10.1371/journal.pone.0125634. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yuan XP, Dong M, Li X, Zhou JP. GRP78 promotes the invasion of pancreatic cancer cells by FAK and JNK. Mol Cell Biochem. 2015;398(1–2):55–62. doi: 10.1007/s11010-014-2204-2. [DOI] [PubMed] [Google Scholar]
- 19.Feng X, Lv W, Wang S, He Q. miR495 enhances the efficacy of radiotherapy by targeting GRP78 to regulate EMT in nasopharyngeal carcinoma cells. Oncol Rep. 2018;40(3):1223–1232. doi: 10.3892/or.2018.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu YF, Liu X, Gao M, Zhang YN, Liu J. Endoplasmic reticulum stress induces autophagy and apoptosis while inhibiting proliferation and drug resistance in multiple myeloma through the PI3K/Akt/mTOR signaling pathway. Oncotarget. 2017;8(37):61093–61106. doi: 10.18632/oncotarget.17862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Zhang B, Lv W, Lai C, Chen Z, Wang R, et al. Triptolide inhibits cell growth and GRP78 protein expression but induces cell apoptosis in original and radioresistant NPC cells. Oncotarget. 2016;7(31):49588–49596. doi: 10.18632/oncotarget.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He B, Luo B, Chen Q, Zhang L. Cigarette smoke extract induces the expression of GRP78 in A549 cells via the p38/MAPK pathway. Mol Med Rep. 2013;8(6):1683–1688. doi: 10.3892/mmr.2013.1724. [DOI] [PubMed] [Google Scholar]
- 23.Yu T, Guo Z, Fan H, Song J, Liu Y, Gao Z, et al. Cancer-associated fibroblasts promote non-small cell lung cancer cell invasion by upregulation of glucose-regulated protein 78 (GRP78) expression in an integrated bionic microfluidic device. Oncotarget. 2016;7(18):25593–25603. doi: 10.18632/oncotarget.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, Guo W, Yang S, Zhu X, Xiang J, Li H. Serum GRP78 as a tumor marker and its prognostic significance in non-small cell lung cancers: a retrospective study. Dis Markers. 2015;2015:814670. doi: 10.1155/2015/814670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soto-Pantoja DR, Wilson AS, Clear KY, Westwood B, Triozzi PL, Cook KL. Unfolded protein response signaling impacts macrophage polarity to modulate breast cancer cell clearance and melanoma immune checkpoint therapy responsiveness. Oncotarget. 2017;8(46):80545–80559. doi: 10.18632/oncotarget.19849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Li Z, Ding G, La X, Yang P, Li Z. GRP78 plays an integral role in tumor cell inflammation-related migration induced by M2 macrophages. Cell Signal. 2017;37:136–148. doi: 10.1016/j.cellsig.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 27.de-Freitas-Junior JCM, Carvalho S, Dias AM, Oliveira P, Cabral J, Seruca R, et al. Insulin/IGF-I signaling pathways enhances tumor cell invasion through bisecting GlcNAc N-glycans modulation an interplay with E-cadherin. PLoS ONE. 2013;8(11):e81579. doi: 10.1371/journal.pone.0081579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denduluri SK, Idowu O, Wang Z, Liao Z, Yan Z, Mohammed MK, et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2015;2(1):13–25. doi: 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spadaro O, Camell CD, Bosurgi L, Nguyen KY, Youm YH, Rothlin CV, et al. IGF1 shapes macrophage activation in response to immunometabolic challenge. Cell Rep. 2017;19(2):225–234. doi: 10.1016/j.celrep.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batista-Silva LR, Rodrigues LS, Vivarini Ade C, Costa Fda M, Mattos KA, Costa MR, et al. Mycobacterium leprae-induced Insulin-like Growth Factor I attenuates antimicrobial mechanisms, promoting bacterial survival in macrophages. Sci Rep. 2016;6:27632. doi: 10.1038/srep27632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y, Chen C, Chen J, Zhan R, Zhang Q, Xu X, et al. Cell surface GRP78 facilitates hepatoma cells proliferation and migration by activating IGF-IR. Cell Signal. 2017;35:154–162. doi: 10.1016/j.cellsig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffenbach KT, Pong M, Morgan TE, Wang H, Ott K, Zhou B, et al. GRP78/BiP is a novel downstream target of IGF-1 receptor mediated signaling. J Cell Physiol. 2012;227(12):3803–3811. doi: 10.1002/jcp.24090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lev A, Lulla AR, Wagner J, Ralff MD, Kiehl JB, Zhou Y, et al. Anti-pancreatic cancer activity of ONC212 involves the unfolded protein response (UPR) and is reduced by IGF1-R and GRP78/BIP. Oncotarget. 2017;8(47):81776–81793. doi: 10.18632/oncotarget.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang YB, Tang H, Chen ZB, Zeng LJ, Wu JG, Yang W, et al. Downregulated SOCS1 expression activates the JAK1/STAT1 pathway and promotes polarization of macrophages into M1 type. Mol Med Rep. 2017;16(5):6405–6411. doi: 10.3892/mmr.2017.7384. [DOI] [PubMed] [Google Scholar]
- 35.Malyshev I, Malyshev Y. Current concept and update of the macrophage plasticity concept: intracellular mechanisms of reprogramming and M3 macrophage "switch" phenotype. Biomed Res Int. 2015;2015:341308. doi: 10.1155/2015/341308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thon M, Hosoi T, Ozawa K. Insulin enhanced leptin-induced STAT3 signaling by inducing GRP78. Sci Rep. 2016;6:34312. doi: 10.1038/srep34312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun S, Yun CW, Lee JH, Kim S, Lee SH. Cripto enhances proliferation and survival of mesenchymal stem cells by up-regulating JAK2/STAT3 pathway in a GRP78-dependent manner. Biomol Ther. 2018;26(5):464–473. doi: 10.4062/biomolther.2017.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett JP, Minogue AM, Falvey A, Lynch MA. Involvement of IGF-1 and Akt in M1/M2 activation state in bone marrow-derived macrophages. Exp Cell Res. 2015;335(2):258–268. doi: 10.1016/j.yexcr.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Zhang Y, Yao G, Gao J, Yang B, Zhao Y, et al. M2-polarized macrophages promote metastatic behavior of Lewis lung carcinoma cells by inducing vascular endothelial growth factor-C expression. Clinics. 2012;67(8):901–906. doi: 10.6061/clinics/2012(08)08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu X, Shi H, Qi Y, Zhang W, Dong P. M2 polarized macrophages induced by CSE promote proliferation, migration, and invasion of alveolar basal epithelial cells. Int J Immunopharmacol. 2015;28(1):666–674. doi: 10.1016/j.intimp.2015.07.033. [DOI] [PubMed] [Google Scholar]
- 41.Dai F, Liu L, Che G, Yu N, Pu Q, Zhang S, et al. The number and microlocalization of tumor-associated immune cells are associated with patient's survival time in non-small cell lung cancer. BMC Cancer. 2010;10:220. doi: 10.1186/1471-2407-10-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DW, Min HS, Lee KH, Kim YJ, Oh DY, Jeon YK, et al. High tumour islet macrophage infiltration correlates with improved patient survival but not with EGFR mutations, gene copy number or protein expression in resected non-small cell lung cancer. Br J Cancer. 2008;98(6):1118–1124. doi: 10.1038/sj.bjc.6604256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J, Liu L, Che G, Yu N, Dai F, You Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer. 2010;10:112. doi: 10.1186/1471-2407-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohri CM, Shikotra A, Green RH, Waller DA, Bradding P. Macrophages within NSCLC tumour islets are predominantly of a cytotoxic M1 phenotype associated with extended survival. Eur Respir J. 2009;33(1):118–126. doi: 10.1183/09031936.00065708. [DOI] [PubMed] [Google Scholar]
- 45.Chung FT, Lee KY, Wang CW, Heh CC, Chan YF, Chen HW, et al. Tumor-associated macrophages correlate with response to epidermal growth factor receptor-tyrosine kinase inhibitors in advanced non-small cell lung cancer. Int J Cancer. 2012;131(3):E227–E235. doi: 10.1002/ijc.27403. [DOI] [PubMed] [Google Scholar]
- 46.Ohtaki Y, Ishii G, Nagai K, Ashimine S, Kuwata T, Hishida T, et al. Stromal macrophage expressing CD204 is associated with tumor aggressiveness in lung adenocarcinoma. J Thoracic Oncol. 2010;5(10):1507–1515. doi: 10.1097/JTO.0b013e3181eba692. [DOI] [PubMed] [Google Scholar]
- 47.Wang R, Zhang J, Chen S, Lu M, Luo X, Yao S, et al. Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung cancer (Amsterdam, Netherlands) 2011;74(2):188–196. doi: 10.1016/j.lungcan.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Zhang L, Zhao Y, Li H, Xiao H, Fu R, et al. Cell-surface GRP78 facilitates colorectal cancer cell migration and invasion. Int J Biochem Cell Biol. 2013;45(5):987–994. doi: 10.1016/j.biocel.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Ayaub EA, Tandon K, Padwal M, Imani J, Patel H, Dubey A, et al. IL-6 mediates ER expansion during hyperpolarization of alternatively activated macrophages. Immunol Cell Biol. 2019;97(2):203–217. doi: 10.1111/imcb.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin K, Ma S, Li H, Wu M, Sun Y, Fu M, et al. GRP78 impairs production of lipopolysaccharide-induced cytokines by interaction with CD14. Front Immunol. 2017;8:579. doi: 10.3389/fimmu.2017.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11(11):750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 52.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1–m2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu R, Li X, Gao W, Zhou Y, Wey S, Mitra SK, et al. Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin Cancer Res. 2013;19(24):6802–6811. doi: 10.1158/1078-0432.CCR-13-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen JJ, Yao PL, Yuan A, Hong TM, Shun CT, Kuo ML, et al. Up-regulation of tumor interleukin-8 expression by infiltrating macrophages: its correlation with tumor angiogenesis and patient survival in non-small cell lung cancer. Clin Cancer Res. 2003;9(2):729–737. [PubMed] [Google Scholar]
- 55.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. 2008;66(1):1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Mu X, Shi W, Xu Y, Xu C, Zhao T, Geng B, et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle. 2018;17(4):428–438. doi: 10.1080/15384101.2018.1444305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26(1):78. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Lopez E, Flashner-Abramson E, Shalapour S, Zhong Z, Taniguchi K, Levitzki A, et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene. 2016;35(20):2634–2644. doi: 10.1038/onc.2015.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Not applicable.