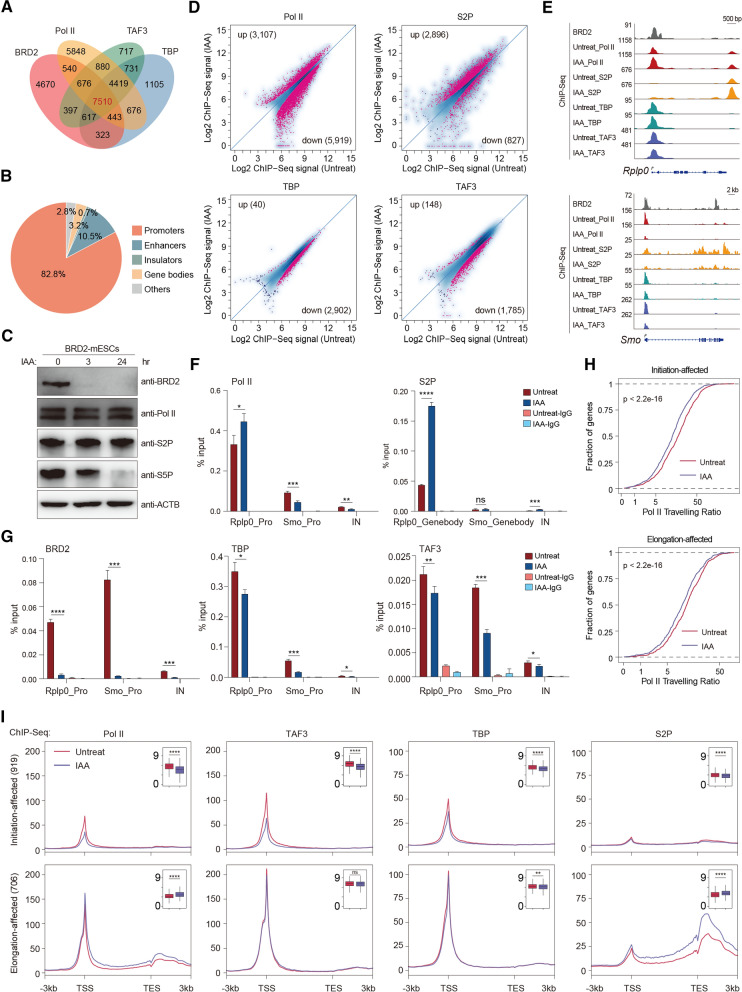
Fig. 1.
BRD2 depletion leads to both Pol II initiation and elongation defects. A Venn diagram displaying the overlapping peaks of the BRD2, Pol II, TAF3, and TBP ChIP-Seq datasets. B The pie chart shows that the overlapping peaks of BRD2, Pol II, TAF3, and TBP ChIP-Seq primarily occupy gene promoters. C Western blot analyses of BRD2, Pol II, S2P Pol II and S5P Pol II protein levels in BRD2 degron mES cells under untreated and IAA-treated conditions. D Scatter plots showing total Pol II (antibody recognized N-terminal of RPB1), S2P (serine 2 phosphorylated RPB1), TBP and TAF3 ChIP-Seq signals at expressed gene promoters and gene bodies (for S2P) before and after BRD2 degradation. Red dots represent the differentially regulated genes identified by DiffBind software (v3.2.2) using DESeq2 as a comparison model (FDR < 0.05). Two replicates were done for each next-generation sequencing experiment. E Genome browser track of BRD2, Pol II, S2P, TBP, and TAF3 ChIP-Seq signals in BRD2 degron mES cells in untreated and 3 h IAA-treated conditions at the Rplp0 and Smo loci. F Pol II ChIP-qPCR analyses of Rplp0 and Smo promoters and intergenic regions in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. Pol II-S2P ChIP-qPCR analyses of the Rplp0 and Smo gene body regions in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. The intergenic region serves as a negative control for ChIP-qPCR. Primers see Table S9. Error bars represent the SD of at least three technical replicates. p values were calculated using Student’s t test (ns: p > 0.05, *p < = 0.05, **p < = 0.01, ***p < = 0.001, ****p < = 0.0001). G BRD2, TBP and TAF3 ChIP-qPCR analyses of Rplp0 and Smo promoters and intergenic regions in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. The qPCR results are represented the same as (F). H The accumulative distribution curve of the Pol ll traveling ratio at initiation and elongation-affected genes upon BRD2 degradation. Initiation-affected genes were defined as genes whose promoters were bound by BRD2, TAF3 and Pol II and showed a decrease in TAF3 signals at promoters identified by DiffBind. Elongation-affected genes were defined as genes whose promoters were bound by BRD2 and Pol II and showed an increase in S2P signals at gene bodies and transcription ending sites identified by DiffBind. Significance was assessed using the Wilcoxon test. I Meta-gene plots of the average Pol II, TAF3, TBP and S2P ChIP-Seq signals at initiation-affected, elongation-affected genes (see methods for more details). Boxplots (insets) displayed the log2 ChIP-Seq signals at ± 100 bp around TSS regions or gene-body regions (Pol II or S2P at elongation-affected genes. Gene body regions were defined as − 1 kb from TSS to transcription termination sites) upon BRD2 depletion. Significance was generated using the ChIP-Seq signals in each gene set under untreat and IAA conditions by the Wilcoxon test (ns p > 0.05, **p < = 0.01, ****p < = 0.0001)