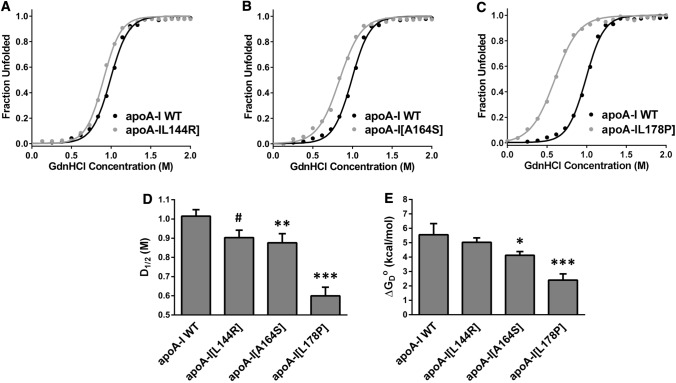
Fig. 4.
Effect of L144R, A164S and L178P mutations on chemical denaturant unfolding of apoA-I. a–c The chemical denaturation profile of each apoA-I mutant is presented in comparison to the WT protein. Solid line indicates the fit of data to Boltzmann sigmoidal model. The y axis has been normalized to correspond to the fraction of the protein in the unfolded state. For normalization, 0 and 1 are defined as the bottom and plateau values, respectively, of the Boltzmann fit. d, e D1/2 and ΔGoD values for the denaturant-induced transitions of WT and mutant apoA-I forms. Values represent the means ± SD (n = 3–4). #p < 0.01; * < 0.05; **p < 0.005; ***p < 0.0001 versus apoA-I WT