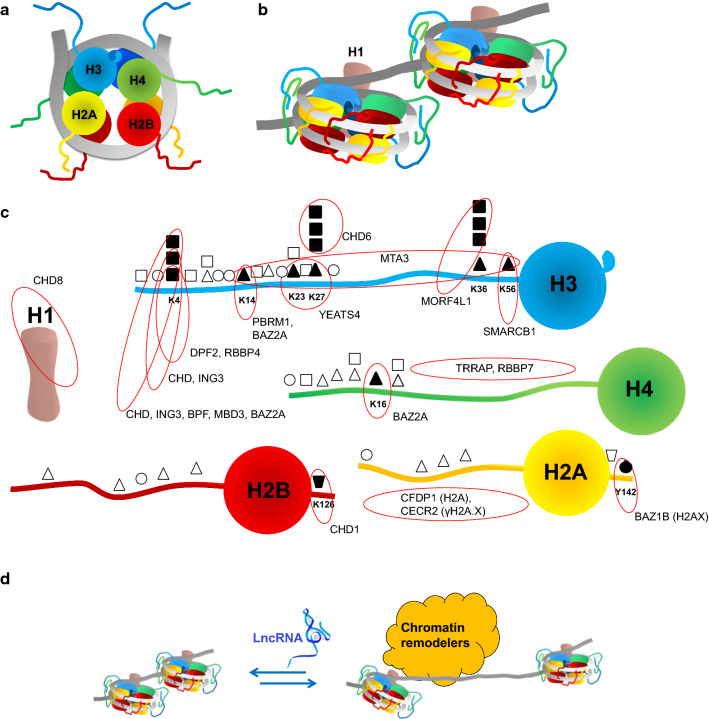
Fig. 1.
Organization of chromatin. Depicted is a DNA helix (in grey) with histone proteins H2A, H2B, H3, H4 and H1 (different colors). a Schematized top view of a nucleosome with tetramer (H3-H4)2 and two dimers (H2A-H2B). b Nucleosomes side view with histone H1 stabilizing the DNA linker. Methylated histone tails have stronger DNA interactions and then acetylated tails. c Presented are common histones modifications, such as methylation (squares), acetylation (triangles), phosphorylation (circles) and ubiquitination (trapeziums). Histones modifications are frequently recognized by chromatin readers. Modifications that are implicated in chromatin remodeling complexes are depicted by solid symbols and their corresponding readers are annotated (red circles). d LncRNAs (in blue) can either inhibit or stimulate chromatin remodeling via interference with remodeler complexes