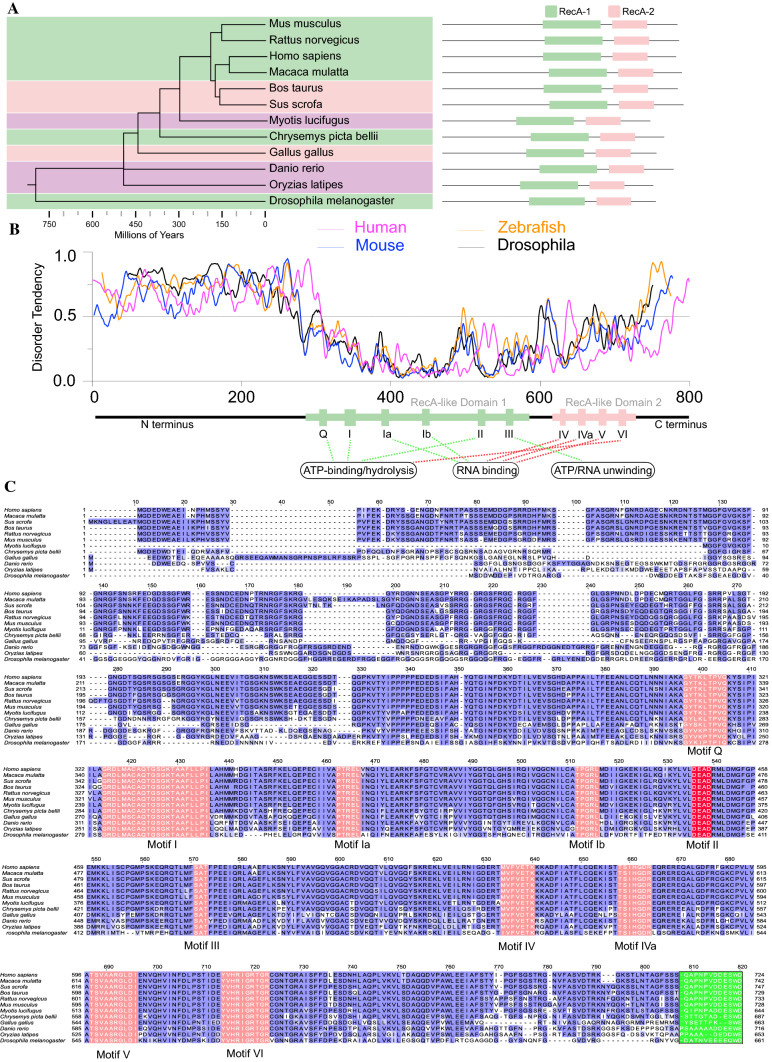
Fig. 1.
Vasa orthologs are evolutionarily conserved and harbor intrinsically disordered regions at both the N-terminus and C-terminus. A The phylogenetic tree of Vasa orthologs from 12 animal species with schematic domain composition illustrated on the right side. Estimated divergence times are obtained from the TimeTree database [102]. B In silico prediction of protein disorder tendency for sequences of Vasa orthologs from mouse (blue), human (red), Drosophila melanogaster (black), and Zebrafish (yellow). Residues above the dotted line are predicted to be disordered by IUPred2A [103]. Conservative regions (DEAD-box helicase domain) are indicated by colored rectangles below. C Multi-sequence alignment of amino acids for Vasa orthologs across 12 metazoan species using Jalview [104]. Amino acids highlighted in red color represent conserved motifs in DEAD-box helicase family (Motifs I–VI) with names labeled, respectively, below. The green color highlights the divergent acidic C-terminal sequence across 12 species