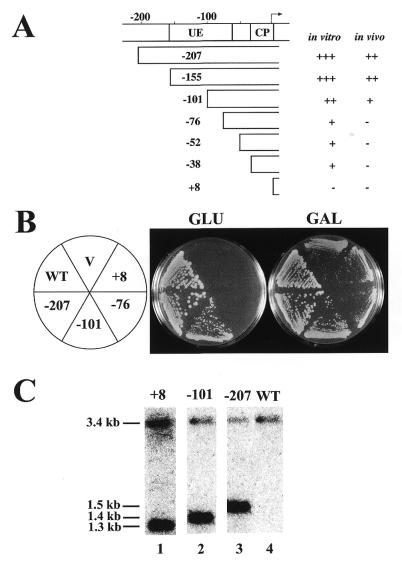
Figure 3.
Deletion analysis of the 35S rDNA promoter using rdnΔΔ strains. (A) The promoter region and various deletion constructs. The names of deletion constructs indicate the last upstream base pair deleted. Numbering is with respect to the transcription start site (+1). Locations of the upstream element (UE) and the core promoter (CP) within the promoter are indicated. A summary of transcription activities of the various promoters in vitro (19) and in vivo [as judged by the extent of rRNA synthesis to allow cell growth (B in this figure and other data not shown) and by rRNA accumulation shown in Fig. 4] is also given. (B) Growth of rdnΔΔ strains on YEPD (Glu) and YEPGal (Gal) plates incubated at 30°C for 5 days. All of these strains carry a helper Pol II plasmid (pNOY353) and deletion derivatives (–101, –76 and +8 as indicated) of a Pol I plasmid (pNOY373, indicated as –207) or a control vector (V). NOY505 (WT) was also included. (C) Southern blot analysis of plasmid copy numbers. DNA was prepared from NOY505 (lane 4, WT), NOY905 (lane 1, +8), NOY909 (lane 2, –101) and NOY908 (lane 3, –207). The DNA preparations were digested with BamHI and EcoRI, subjected to agarose gel electrophoresis and analyzed using a radioactive probe containing LEU2. A picture obtained with a PhosphorImager is shown. The fragment containing the chromosomal LEU2 gene has a calculated size of ∼3.4 kb and that from pNOY373 (in NOY908) has a calculated size of ∼1.5 kb. The sizes of those from the other two plasmids are shorter due to the deletions. Quantification of plasmid copy numbers relative to chromosomal LEU2 gave the following values: pNOY445 (in NOY905), 12; pNOY444 (in NOY909) and pNOY373 (in NOY908), 70–100.