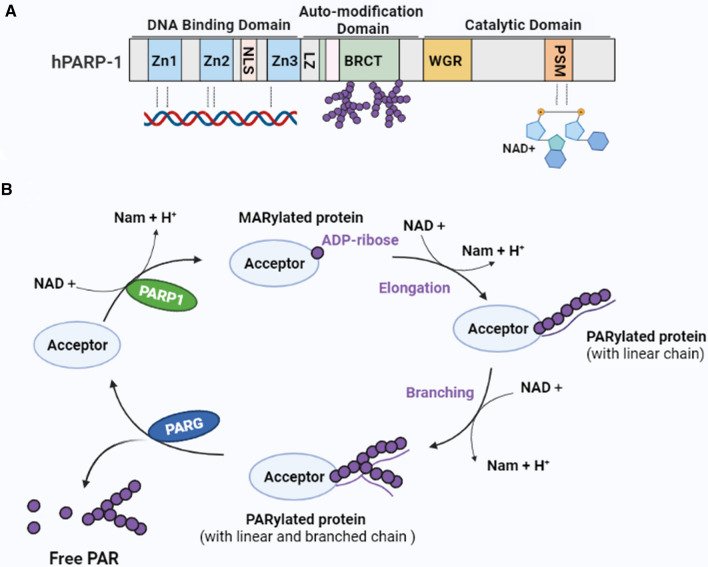
Fig. 2.
Structure of PARP-1 and the production and degradation of PAR. A Structure of PARP-1. PARP-1 includes three structural domains: (1) An N-terminal DNA-binding domain, which contains three zinc-binding domains (Zn1, Zn2, and Zn3) and an NLS. The zinc-binding domains are responsible for PARP-1 detection of damaged DNA. (2) A central automodification domain, which contains a leucine zipper (LZ) motif and a BRCA1 C-terminal (BRCT) phosphopeptide-binding motif for protein–protein interaction. (3) A C-terminal catalytic domain that contains a WGR domain and the “PARP signature” sequence (PSM), which catalyzes PAR synthesis using nicotinamide adenine dinucleotide (NAD +) as a donor of ADP-ribose. B Production and degradation of PAR. PARP-1 uses NAD + as a substrate to generate mono(ADP-ribosyl) and nicotinamide (Nam). Mono(ADP-nucleotide) is covalently attached to the target protein, resulting in MARylated protein. Further elongation and branching of the polymer forms PARylated protein. PARG catalyzes the hydrolysis of PAR to free poly(ADP-ribose) or ADP-ribose units through its endoglycosidic or exoglycosidic activity. PARP-1 poly(ADP-ribose) polymerase 1, PAR poly(ADP-ribose), PARG PAR glycohydrolase